# Title : 12092019-Transformation of ylic133 with the pLL112 plasmid 😔
# Date
12092019-20092019
# Objective
To mark temporarily ylic133 in order to be able to select for diploids after the mating with yEK7a.
# Method
Plasmid transformation using pLL112: pRS416 (URA3 CEN6_ARSH4)
12092019: Incubation of ylic133_2 in 10mL YP+2% Dextrose
13092019: OD measurements @9:30am
OD-10X dilution Titer Dilution factor to OD=0.5 Time ylic133_2 0.259 2.6 5.2 @9:30 2h after the 5x dilution of the dense culture in 10mL YPD
OD-10X dilution Titer Time ylic133_2 0.126 1.3 @11:30 OD-10X dilution Titer Time ylic133_2 0.239 2.4 @13:30 5ul of pLL112 plasmid , which has 2290ng/mL
# Results
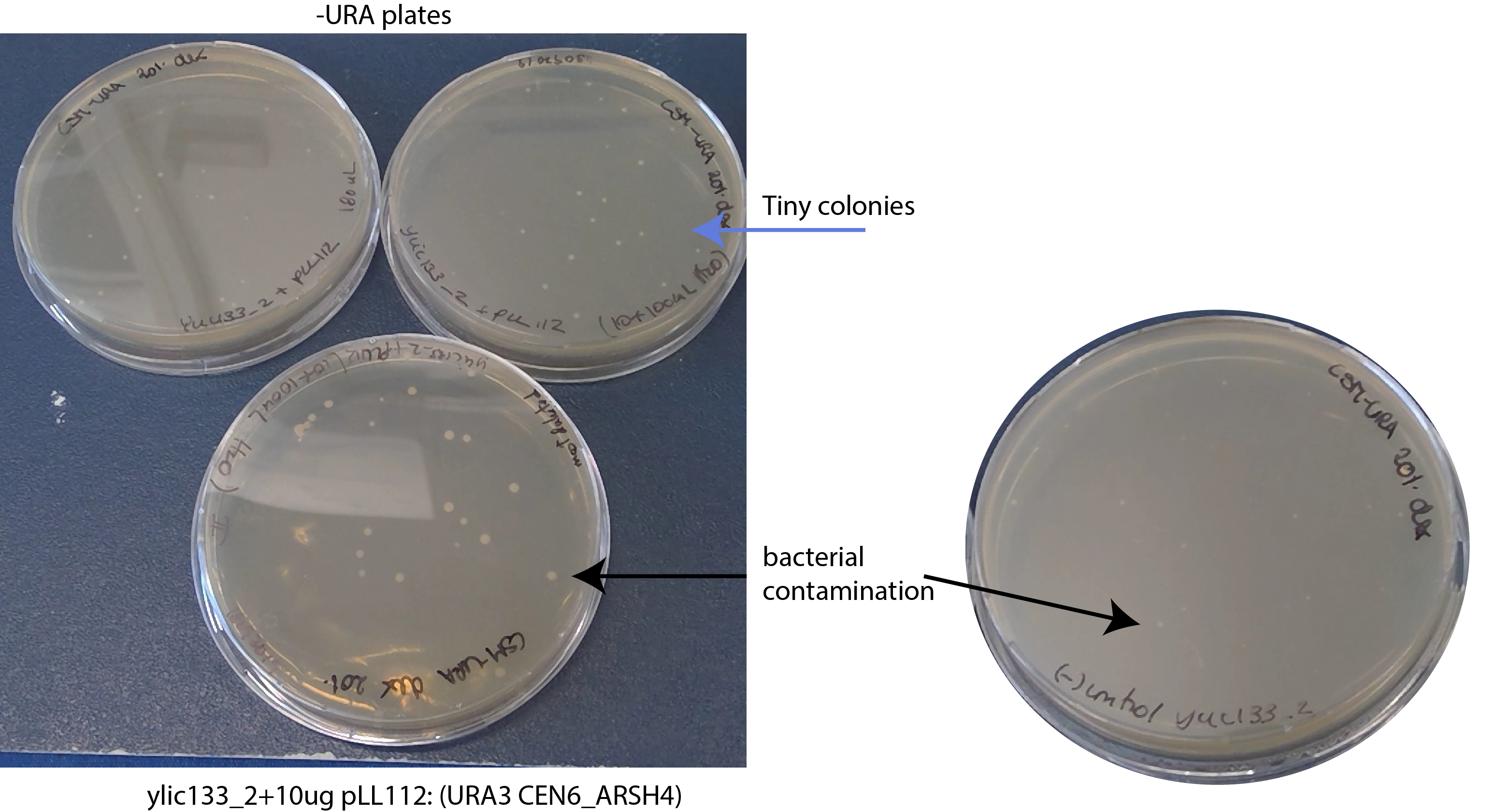
- 17092019- I found bacterial contamination on the plates 😦
- 17092019- The transformants on the -ura plate are extremely tiny colonies
- 17092019 - Incubate ylic133-3 to redo transformation
- 18092019 - The liquid culture was not dense enough to do the experiment.
- 19092019 - I diluted the culture 1000x to do transformation next day.
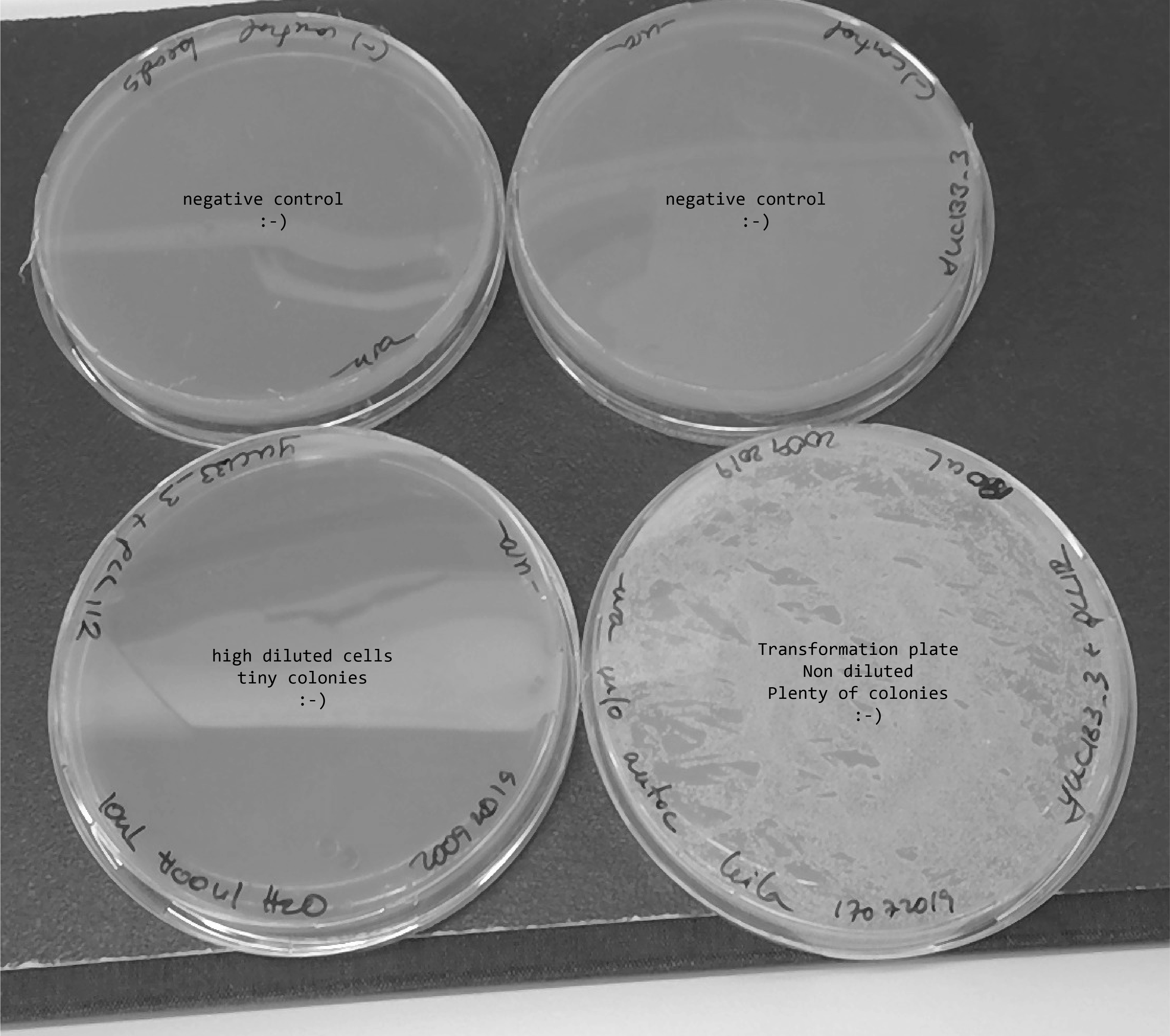
- 20092019 - Transformation with pLL112 (from a miniprep Ramon did the day before)
- pLL112-148ng/ul , I used 10ul for transformation , hence I used 1.5 ug of plasmid
- I plated two positive controls in -ura and all the negative control in -ura.
# Conclusion
- I got very small transformed strains which are going to be named: ylic134=ylic133_3+pLL112 (mating type alpha, ura3+)
- I inoculated transformants colonies into a CSM-URA liquid media and they did not grow , from an overnight culture. 😦
- It seems somehow they loose the plasmid... 😔
- As next step I will transform ylic133 and yll3a as a positive control with pLL112, pLL55 and pLL56, to assess whether is the plasmid or the strain the bottleneck here.