# Title: Literature search
# Goals for the literature search:
- To collect relevant published information about how the prediction of synthetic lethals has been generally carried out .
- To be able to reproduce existing results (if possible) and to have a culture of how other researchers have tackled this question.
# General goal of the project
- Our general goal is to apply supervised machine learning for this task of predicting synthetic lethals, because we have a huge amount of data that we can use for training and testing our methods. Hence, the part of testing if our predictions satisfy the current knowledge is straighforward and the purpose of the a supervised learning algorithm.
# Benefit from the literature search to the application of the supervised machine algorithm
- Informative datasets that has been used for the prediction of synthetic lethality.
- Design of insightful and relevant features to construct the learning.
# Papers related to the prediction in silico of synthetic lethals
# Paper title: "Predicting yeast synthetic lethal genetic interactions using protein domains" 2009
# Goal:
- Predicting SL interactions based on the protein domains a protein pair share.
# Data sources:
- The protein domain data was collected from Pfam (Protein families database)
- Genetic interactions of yeast were downloaded from the Saccharomyces Genome Database (SGD)
# Method:
- Support vector machine (SVM) for two class classification problem, of predicting if certain pair of proteins can be synthetic lethals or not.
- They used LibSVM tool in C to implement the method.
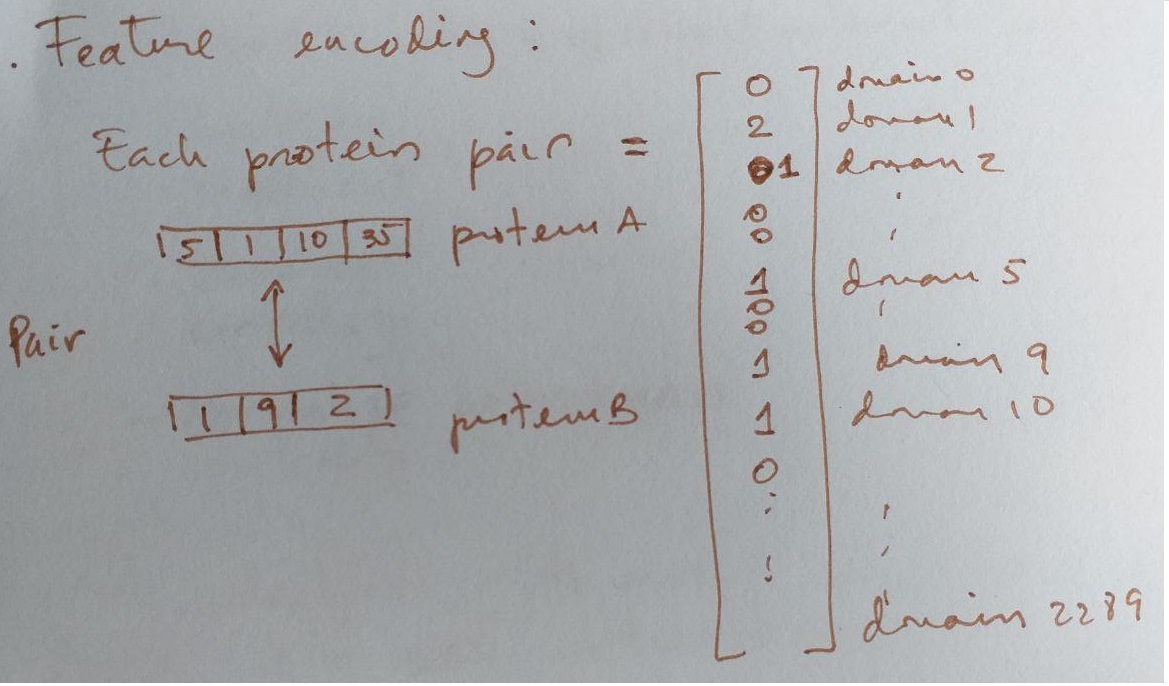
# Feature encoding:
- Use of the protein domains to find correlations with existing synthetic lethals pairs in order to predict new ones.
 {#fig:feature width=50%}
{#fig:feature width=50%}
# Paper Title: "Mining protein networks for synthetic genetic interactions" 2008
# Goal:
- To model correlations between protein interaction network properties and the existence of synthetic lethal interaction.
# Data sources:
- Protein-protein interaction dataset from Reguly et al['Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae'] This dataset contains 3289 proteins and 11334 interactions.
# Method:
- Support vector machine (SVM) for two class classification problem, of predicting if certain pair of proteins can be synthetic lethals or not.
# Feature encoding:
- Use of protein interaction network graph theoretic properties:
Global properties:
-a Degree
-b Clustering coefficient
-c Closeness centrality
-d Normalized betweenness centrality
Local properties:
- the inverse of the shortest distance
- number of mutual neighbors between proteins p and q
- 2Hop S-S and 2Hop S-P
[1]: which exploit the fact that the known synthetic genetic network contains a large number of triangles. 2Hop S-S takes a value 1 if the genes encoding the two proteins p and q share a synthetic lethal partner and 0 otherwise, whereas 2Hop S-P takes a value 1 if there exists a protein r such that r has physical interaction with protein p and the gene corresponding to protein r has a SSL interaction with protein q or vice versa.
# Paper title: "Predicting genetic interactions with random walks on biological networks"
# Goal
The application of randon walks to accurately predict pairwise interactions.
# Data sources
- Gene Ontology
- protein-protein interaction dataset
- Data on synthetic sick and synthetic lethal interactions (BioGrid, collins et al, Davierwala et al, Tong et al)
# Method
- Random walks to determine the strength of the proximity between two nodes.
- Decision Tree classifier
# Feature enconding
- The proximity matrices are combined with the genetic interaction data during the procedure for measuring the topological relatedness between two genes. This generates a large vector for gene pair that are introduced as feature for the decision tree.