# Title : 20200810-SATAY for $\Delta$bem2, $\Delta$nrp1 and WT
# Date
10082020 - 18082020
# Objective
- To test the technique on the mutants and also to have data for the data analysis pipeline from Greg.
# Method
10082020-Preculture in SD-URA+0.2%glucose +2% raffinose at 14:20
- Scrape from the colonies from each background that passed the test (2 per each background)
- Inoculate to a final volume of 20mL and to a final OD of 0.2
11082020
- OD measurement
OD really low around 0.3 for all strains which could be due to the fact that the media did not contain extra adenine.
SO I RESTARTED THE PRECULTURE ON 11082020
- OD measurement
12082020 - Induction in SD-URA+2% gal to a final OD of 0.2 - 50 hours (2 days)
- OD measurement:
| Strain | OD 10x dilution | ReaL OD | Dilution Factor to OD=0.2 | Time of preculture |
|---|---|---|---|---|
| ylic133_1 | 0.659 | 6.6 | ~30 | 20h |
| ylic133_2 | 0.577 | 5.7 | ~30 | 20h |
| ylic135_1 | 0.073 | 0.7 | ~3 | 20h |
| ylic135_2 | 0.126 | 1.26 | ~6 | 20h |
| ylic136_1 | 0.678 | 6.7 | ~30 | 20h |
| ylic136_2 | 0.626 | 6.3 | ~30 | 20h |
Issues :
- The ylic135 strains grows a bit more slower than the rest which is expected due to the phenotype of not having bem2. But the amount of culture I need to add in order to have a final OD of 0.2 in 150mL is 50 mL and I only had 20mL. Therefore the initial OD of the induction will be around 0.1 for this strain.
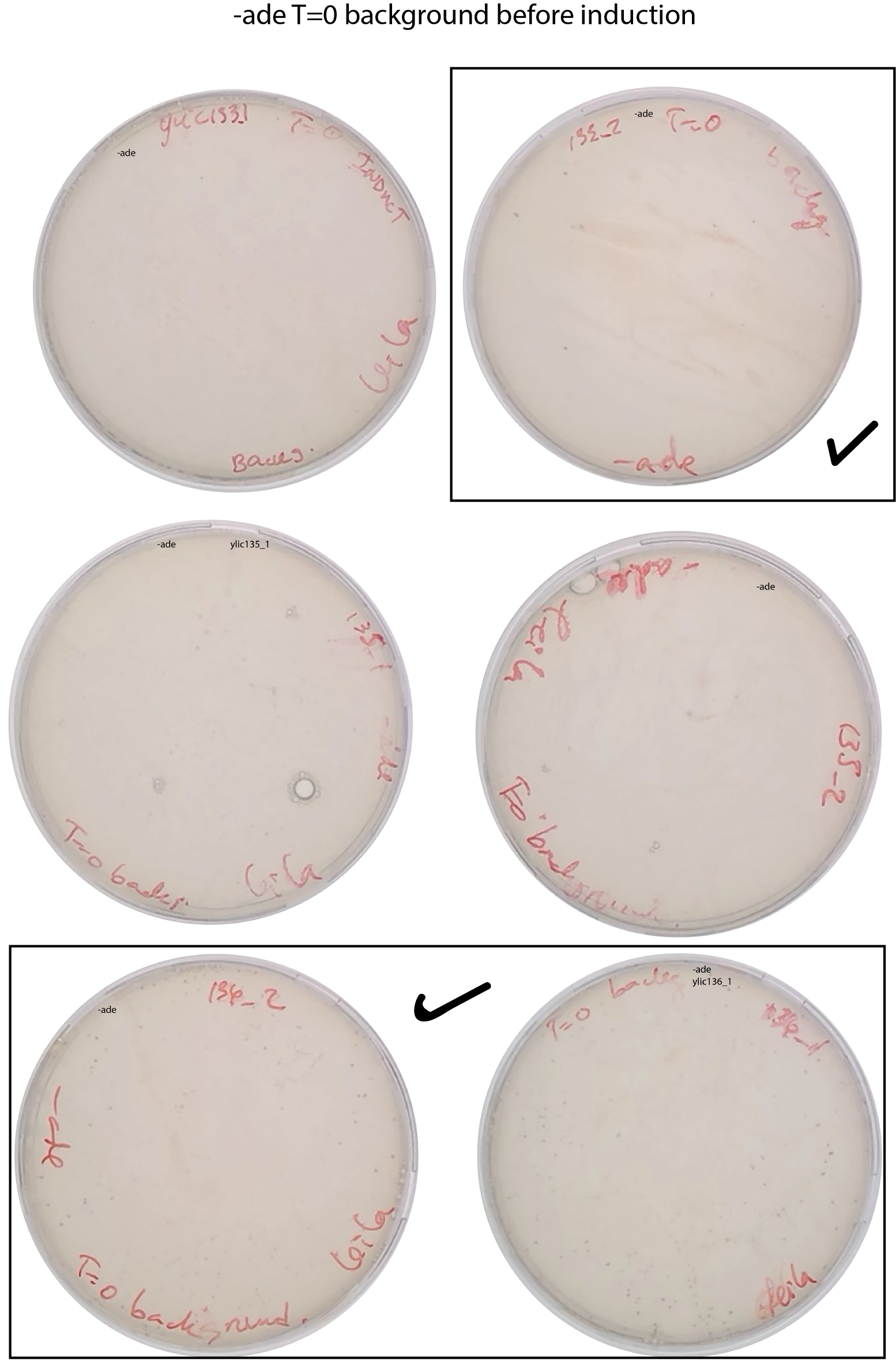
Measure the background (T=0 of induction)
[x] Plate 200ul of the inoculum in SD-ADE+ 2% dextrose (expect 40-80 colonies per 200uL)
[x] Dilute 1000x and spread 200ul in SD-URA
[x] Prepare 3 flasks of 3L of SD-ADE media for the reseed of three strains
- Per flask of 3L , add: 2,4L MiliQ to autoclave, dissolve 2.34g of -ade drop out + 20.7g of YNB (005) and filter sterilize it and add it after autoclaving the MiliQ. Add 300mL of 20% dextrose after autoclaving.
13082020
[x] Plating in SD-ADE to check induction after T=22h and SD-URA 1000x dilution.
[x] Check OD (should be around 4-5) at 10:45am
| Strain | OD 10x dilution | ReaL OD | Time |
|---|---|---|---|
| ylic133_1 | 0.472 | 4.7 | 24h |
| ylic133_2 | 1.17 | 11.7 | 24h |
| ylic135_1 | 0.223 | 2.23 | 24h |
| ylic135_2 | 0.217 | 2.17 | 24h |
| ylic136_1 | 1.17 | 11.7 | 24h |
| ylic136_2 | 1.13 | 11.3 | 24h |
Strains ylic133_2 and ylic136 have a very high OD
The cell culture of ylic133_1 have turned pink...
Still the background at T=0 can not be clearly determined.
14082020 - End of induction/Reseed at 14:00
- Choose the 3 strains to continue for the reseed that has the least number of ADE+ cells at T=0 and the highest number of ADE+ after induction.
- It seems that ylic133_2, ylic136_1 and ylic136_2 are the ones that have the least background at T=0 before induction.
- Choose the 3 strains to continue for the reseed that has the least number of ADE+ cells at T=0 and the highest number of ADE+ after induction.
Background check at 9:30am
| Strain | ade+ clones/mL | ura+ clones/mL | Time |
|---|---|---|---|
| ylic133_1 | < 5 | ~100* 5* E3 = 5*E5 | 0h Induction |
| ylic133_2 | ~30*5 = 150 | 5 *E5 | 0h Induction |
| ylic135_1 | - | - | 0h Induction |
| ylic135_2 | - | - | 0h Induction |
| ylic136_1 | ~ 80*5=400 | 5 *E5 | 0h Induction |
| ylic136_2 | ~ 80*5=400 | 5 *E5 | 0h Induction |
- Summary of the background
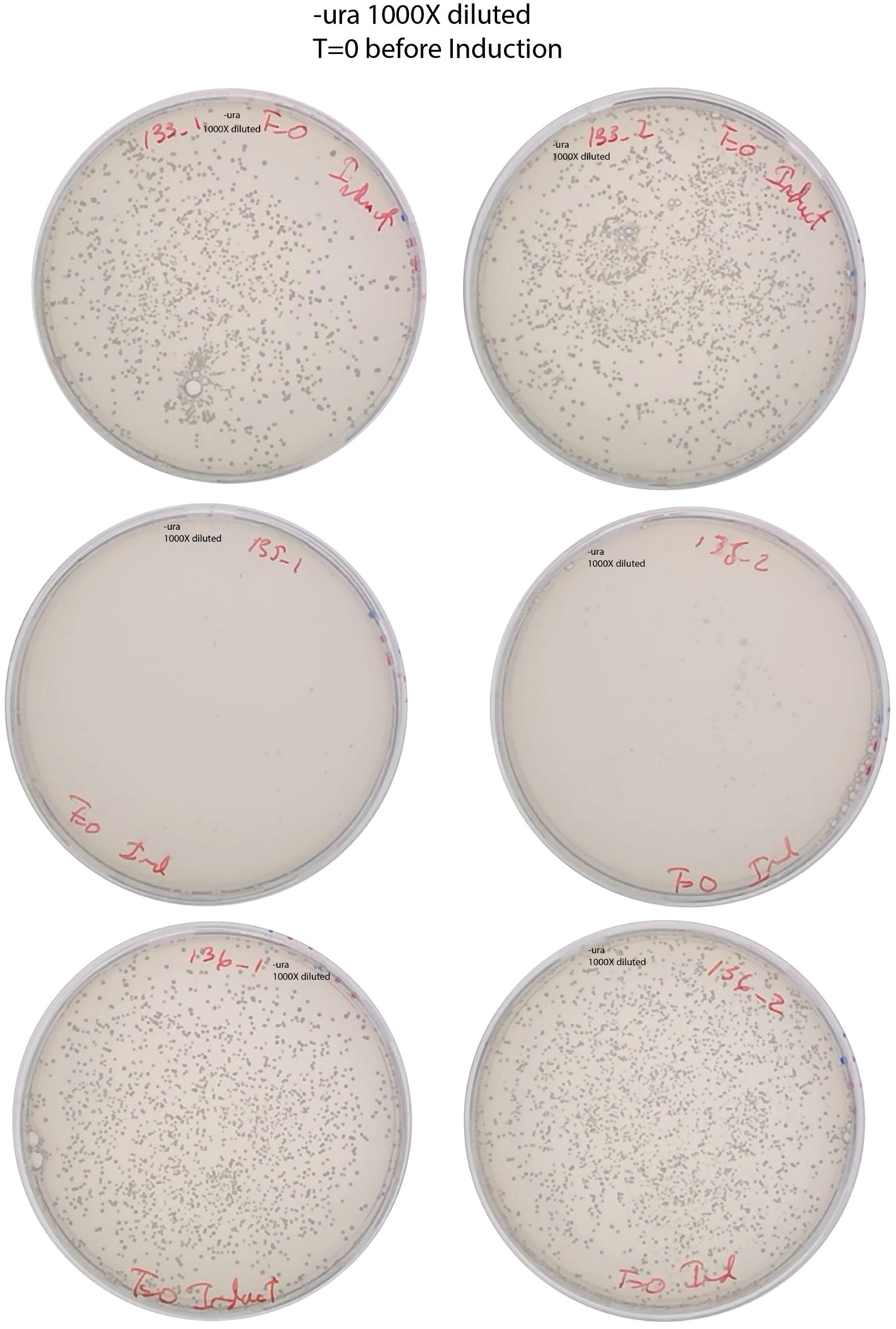
- [x] Plate 200ul from the induction culture in -ade and 1000X in -ura to know the number of ade+ cells before reseeding. At T=51h of induction, at 13:00.
- [x] OD measurement of the samples at T=51h of induction and T$_r$=0
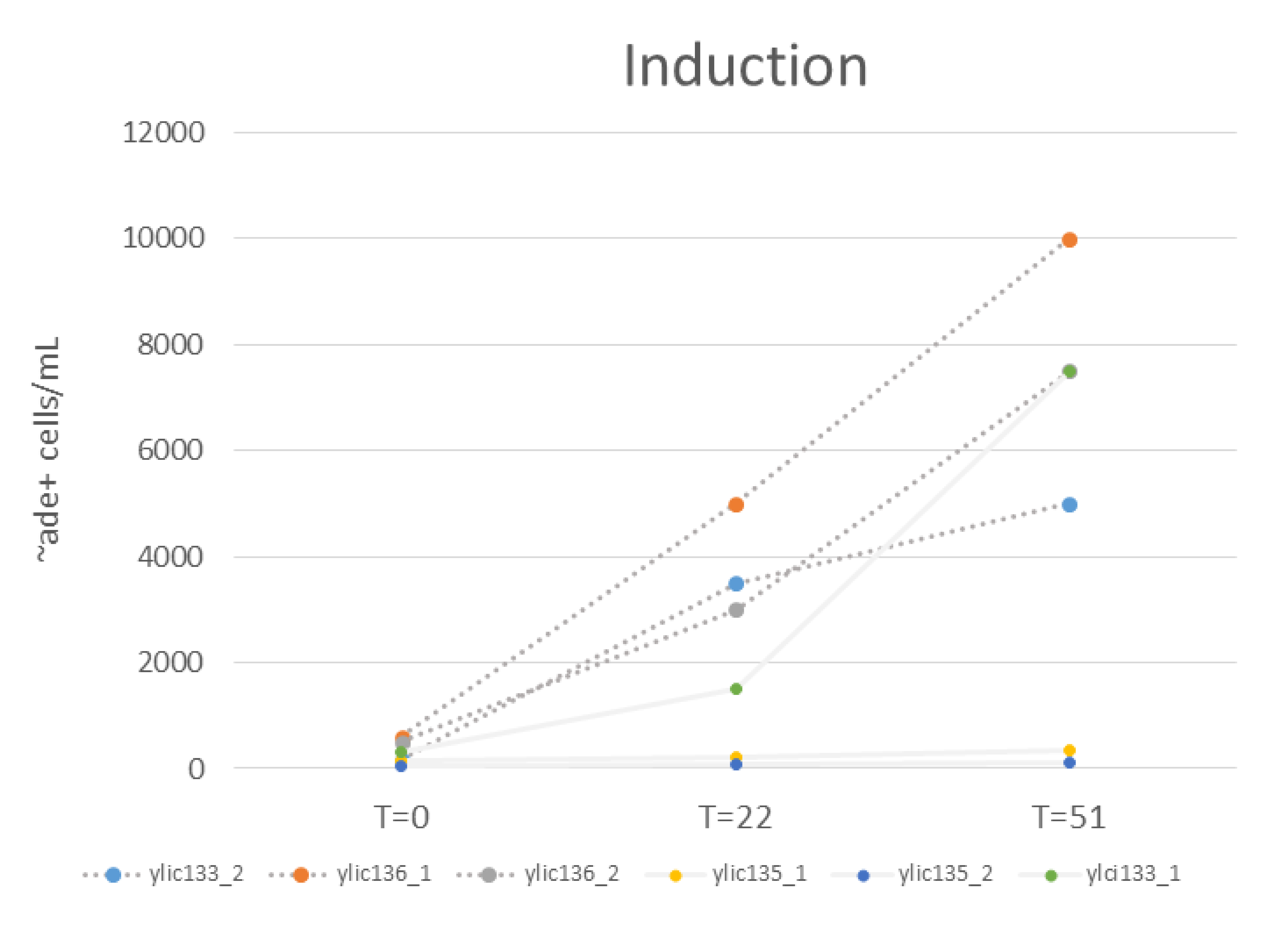
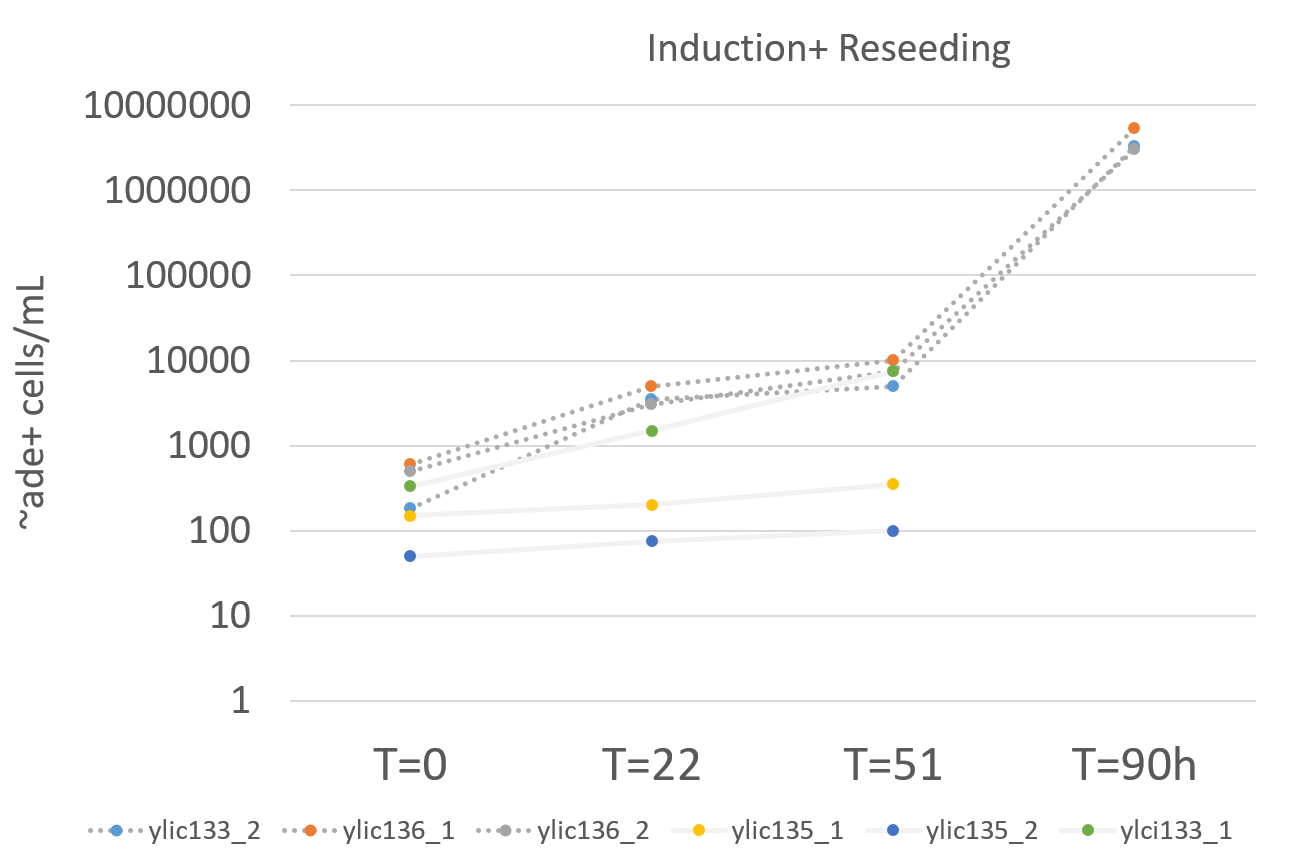
- Summary from the Induction
| Strain | OD start T=0 | OD T=24h | OD stop T=51h | Time-Induction | ADE+/ml-start | ADE+/ml-24H | ADE+/ml-stop |
|---|---|---|---|---|---|---|---|
| ylic133_1 | 0.2 | 4.7 | 5.4 | 51h | 330 | 500 | 7500 |
| ylic133_2 | 0.2 | 11.7 | 13.9 | 51h | 180 | 3500 | 5000 |
| ylic135_1 | 0.1 | 2.23 | 6.5 | 51h | 150 | 200 | 350 |
| ylic135_2 | 0.1 | 2.17 | 6.3 | 51h | 50 | 75 | 100 |
| ylic136_1 | 0.2 | 11.7 | 13.8 | 51h | 600 | 5000 | 10000 |
| ylic136_2 | 0.2 | 11.3 | 12.9 | 51h | 500 | 3000 | 7500 |
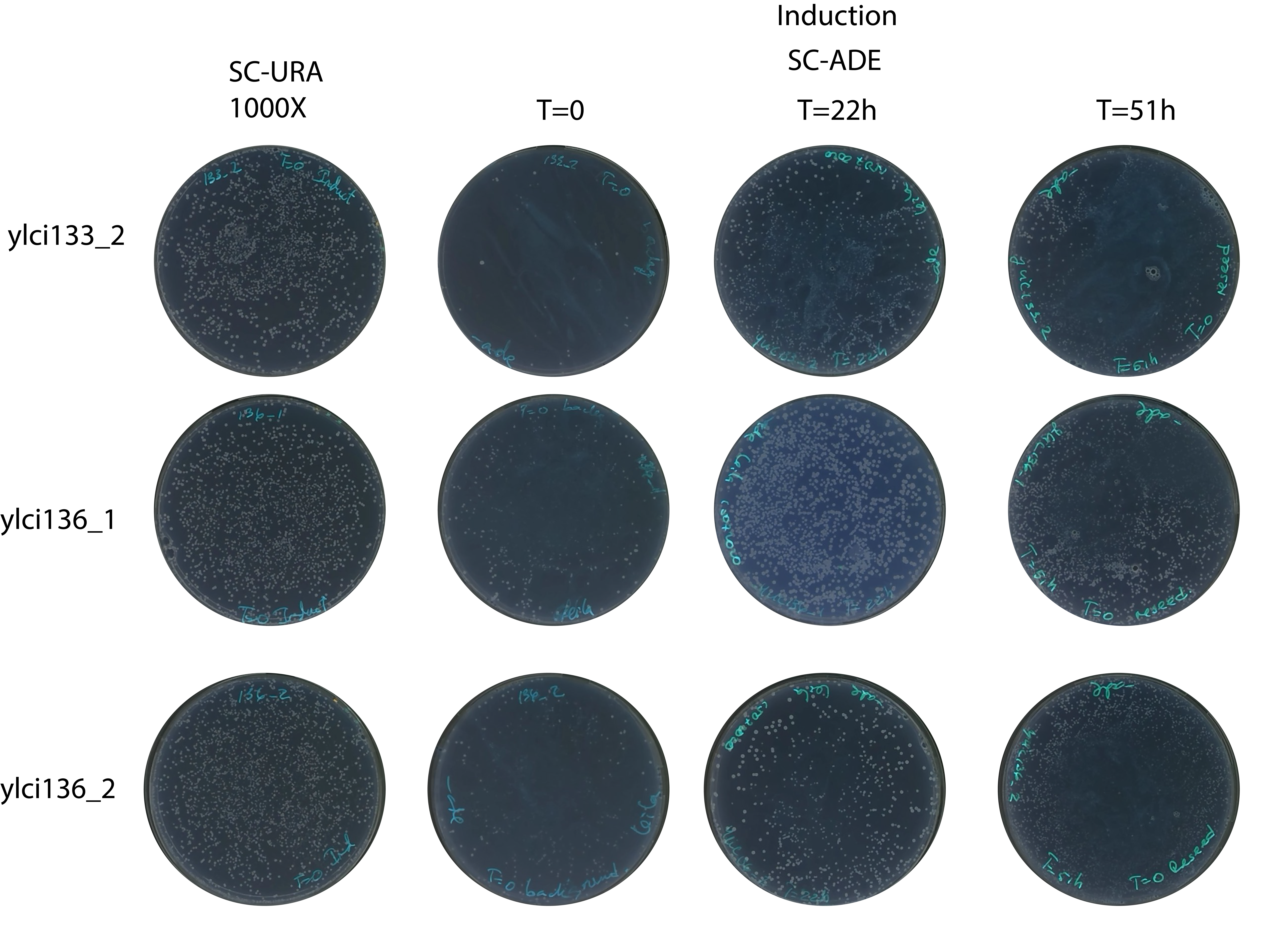
[x] Reseed at 13:30
17082020
[x] Check ADE+ cells during induction. Counting colonies from each plate.
- Image J protocol:
- select image of interest: circle tool and edit-> clear outside
- Image -> Type -> 16bits
- Make sure to capture the maximum number of right colonies: Image -> Adjust -> Threshold . Make sure the colonies are all red.
- Process -> Make binary-> Watershed
- Analyse -> Analyse Particles
- Image J protocol:
18082020
[x] OD measurement for T=92h after reseeding at 12:00
[x] Plate 200ul with 1000X dilution in SD-Ade , and 200ul with 40000X dilution in SD-ura to estimate the growth of ade+cells compared with the T=0 of reseeding. Expect that the ade+ cells have grown by a factor of ~1000x.
End of reseeding
[x] Harvest of the cell culture.
- 15ml of solid pellet, frozen in -80C. The pellet is pink (?)

- 15ml of solid pellet, frozen in -80C. The pellet is pink (?)
| Strain | OD START | OD STOP | Time- Reseeding | ADE+/ml-stop |
|---|---|---|---|---|
| ylic133_2 | 0.25 | 8.3 | 90h | 3295000 |
| ylic136_1 | 0.27 | 10.2 | 90h | 5350000 |
| ylic136_2 | 0.18 | 9.5 | 90h | 3070000 |
# Results
For the strain ylic135 it did not work, because it is very miserable to culture with others. It grows much more slower , so maybe growing it more and with less volume , for next time. Also ylic135 gives a relatively high background compared with the growth in -URA.
The way of computing the number of colonies in highly dense plates is pretty inaccurate with the best tool to do it which is ImageJ. So I think that is why I get like an order of magnitude lower than the number of colonies expected by Benoit.
The pellet from the reseed culture was pink , which could mean that there were many cells ade- that did not have any transposon event. Hence I suspect the sequencing/transposon coverage will be not optimal.
The number of ADE+ cells after reseeding is around 1000X times higher than at then of the induction which is what was expected 😃
# Next steps
- DNA sequencing protocol
- Repeat the SATAY for ylic135 from the beginning
# Conclusion
- First round of SATAY 😃 with WT and dnrp1 strains.