# Title : Preparation for PCR check of BEM2 presence and absence in strain ylic135
# Date
07062020-
# Objective
- [ ] Repeat the test done on beginning of May to see if I get something usable...
- [ ] Check the presence of the leu2 marker in the position of bem2 gene , and checking the bem2 absence in the mutants strains.
# Method
Colony PCR with freshly grown cells from glycerol stocks.
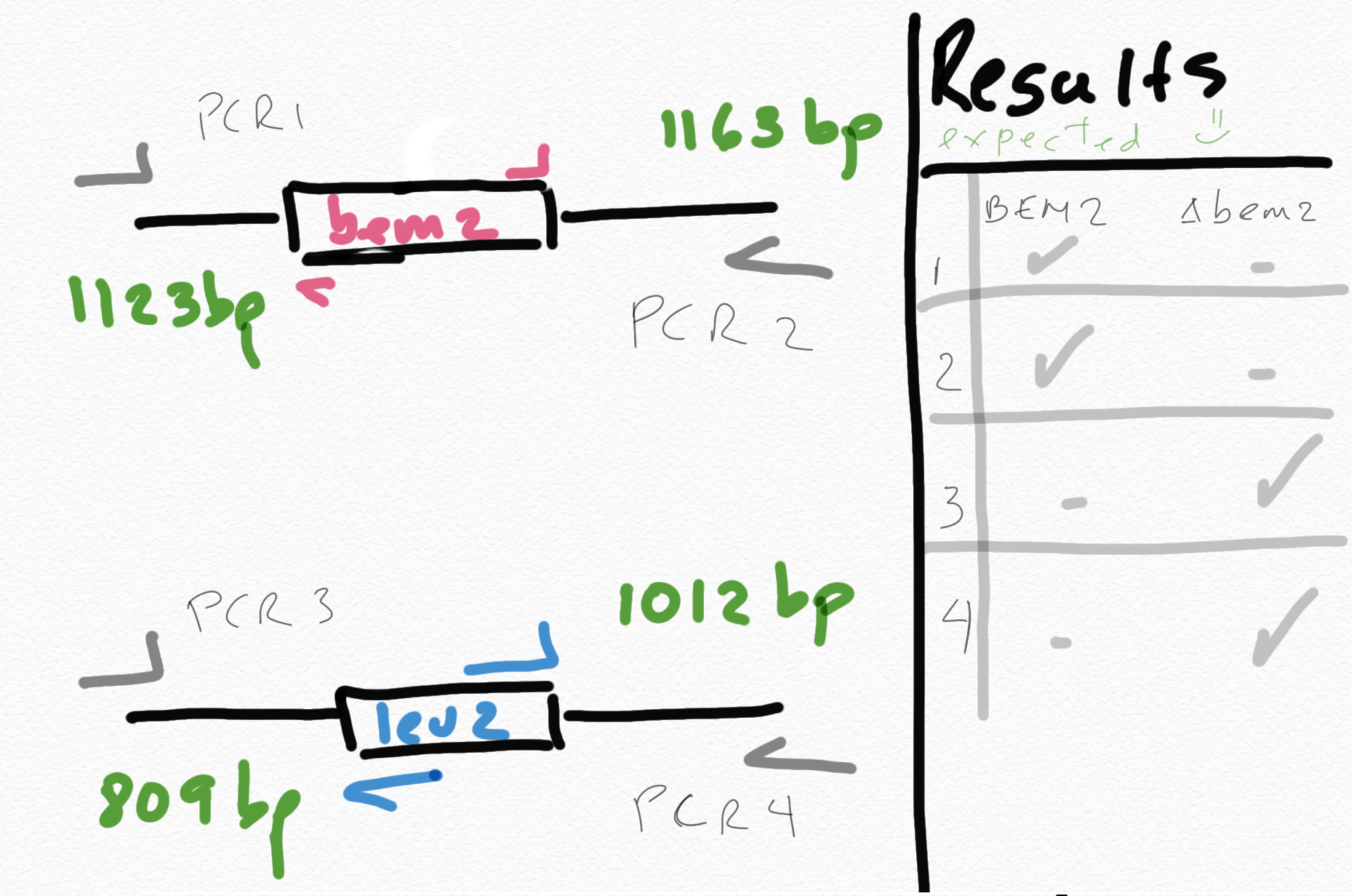 {#fig:sketch-pcr}
{#fig:sketch-pcr}06072020
- 10:00am Plate all glycerol stocks of ylic135 in -leu2 plates , and in YPD
07062020
- No growth yet in -leu2 plates .
- Pink colonies in YPD
08072020
- Colony PCR at 10:30am - 12:30pm.
- Protocol "Leila"
- 5ul Template
- Primers 49 y 50
- I added 1ul of DNA template from yll3a as a positive control for Bem2 presence, with primers 47 y 48.
# Important Note!!!!!!!!
THESE PCR WOULD NEVER WORKS BECAUSE THE PRIMER SET TAKEN FOR EACH TEST WAS WRONG!!!!!!!!!!!!
I was taking only the pink or the blues primer set, looking at the picture above, hence I would never get a band and the region tested was completely wrong!!!
The right primer set combination is:
- 41/49 and 50/42 primer set to test the presence of leu2 marker in the mutants. So 2 PCR per strain.
- 41/47 and 48/42 primer set to test the presence of BEM2 in the WT.
For 5 biological replicates there are 10 PCRs to test the leu2::bem2 construct plus 2 PCRs for yll3a positive control. So 12 PCRs per test.
[x] 08072020- Plate all biological replicates of ylic135 in -leu2+6xade plates and YPD.
[x] 10072020- PCRs with the right primer set at 10:30am
- Total : 24 PCRs
- 12 positive controls for leu2:bem2 locus and BEM2 in WT
- primer set 41/49 , 42/50 for the mutants and 41/47 and 42/48 for WT
- 12 negative controls for not having BEM2 in leu2 locus and not having leu2 marker in BEM2 locus in WT.
- primer set 41/47 and 42/48 for the mutants and 41/49 and 42/50 for the WT , yll3a genomic DNA.
[x] Gel 110V 25 mins
# Results
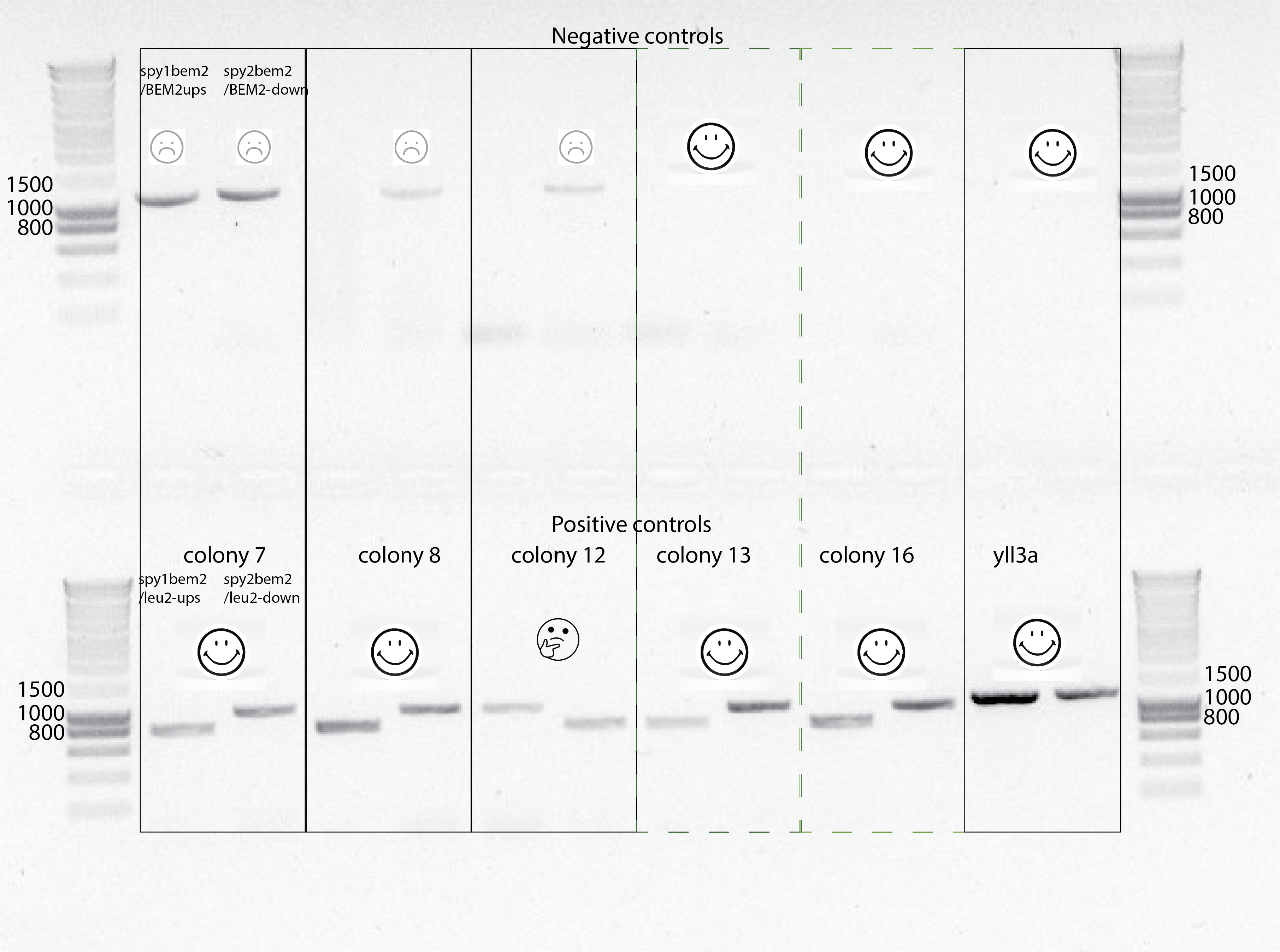
- Finally got some bands due to the use of the right primers!!
- It seems the right strains are the slowest ones , colony 13 and 16. All of the rest , it seems they also have BEM2 there (weird..) and the leu2 marker in the right position.
- It seems colony 8, 12 and 7 are diploids (?)
# Conclusion
- colony 13 and 16 seems to be the right one that has leu2::bem2 and no BEM2 present.
- They are also the slowest one from the Biotek measurement of population growth rates.
# Next Steps
- Transform them with the plasmid pBK549 to do SATAY on them, hopefully also send the data as Enzo for sequencing.