# Title: 29072019-SATAY 1st attempt with Byk832-T 🙏 😕 ❌
# Date
Tuesday, 29072019 - 31072019
# Objective
To learn about the whole technique and detect the main issues and bottlenecks when used it for the target strain.
# Method
Streak the chosen Byk832 transformed cells that display full growth in -URA and medium growth in -ade, in -ura.
Pre-culture step - 29072019 🕝 (14:30) 😊
Scrape cells from SD-URA, inoculate 20-30mL SD-URA 0.2% glucose +2% Raffinose at OD600=0.16-0.24 (OD=0.06 works as well)
it is more than one cell, just take with a pipette tip many cells and put them in a eppi with 1ml MiliQ
Measure the OD of a 100x diluted stock (10ul of diluted cells + 990ml MiliQ)
| Strain | OD (100x diluted) | |---|---| |Byk832T_1 | 0.15 | |Byk832T_2 | 0.24 | |Byk832T_3 | 0.20 |Dilution to OD=0,2 to a final volume of 20mL
Enzo did three flasks more , as technical replicates.
Incubate at 30C, 140-160 rpm, to OD=4-6 (around 20h)
Started at 🕝 (14:30) 😊
It is recommended to start with 3-4 precultures and later chose which to reseed in SD-ADE 2% Glucose.
# Benoit Data
Strain Preculture OD start OD stop Time strain # 1 0.234 4.33 20h strain # 2 0.233 4.02 20h strain # 3 0.235 3.85 20h strain # 4 0.227 4.04 20h strain # 5 0.229 3.94 20h strain # 6 0.235 4.07 20h # Our data
Strain (2 technical replicates) Preculture OD start (29072019-🕝 -> 14:30) OD stop (~2X higher
than Benoit data)Time Byk832T_1(Leila) 0.19 ~ 12.6 19h Byk832T_2 (Leila) 0.14 ~11.2 19h Byk832T_3(Leila) 0.17 ~ 10.8 19h Byk832T_1(Enzo) 0.19 ~ 8 19h Byk832T_2(Enzo) 0.14 ~ 10 19h Byk832T_3(Enzo) 0.17 ~ 9.7 19h Induction
Inoculate 200ml SD-URA 2% Galactose at OD600=0.2 from the saturated preculture.
We choose base on the OD values Byk832T_1(Enzo),Byk832T_2(Enzo), Byk832T_3(Enzo), and Byk832T_3(Leila) forthe induction in 150mL of media
Incubate at 30C , 140 to 160 rpm for 50-52 hours
30072019-🕥 -> 10:30 start T=0 of induction.
To evaluate the background:
- [x] Spread 200ul of this inoculum on SD-ADE
- [x] Dilute the inoculum 1000x, spread 200ul on SD-URA and SD complete
score after 2 days:
[x] 😔 Expect ~200-400 ADE+ clones/mL culture (~ 40-80 clones for 200ul), i.e. ~0.01%-0.02% of the total number of cells. These are cells that repaired the ADE2 gene by homologous recombination WITHOUT transposition
The plates next morning had easily more than 200 colonies , which means 5 times more than indicated for Benoit.

Reproduce the following plots, with the chosen technical replicates for induction:
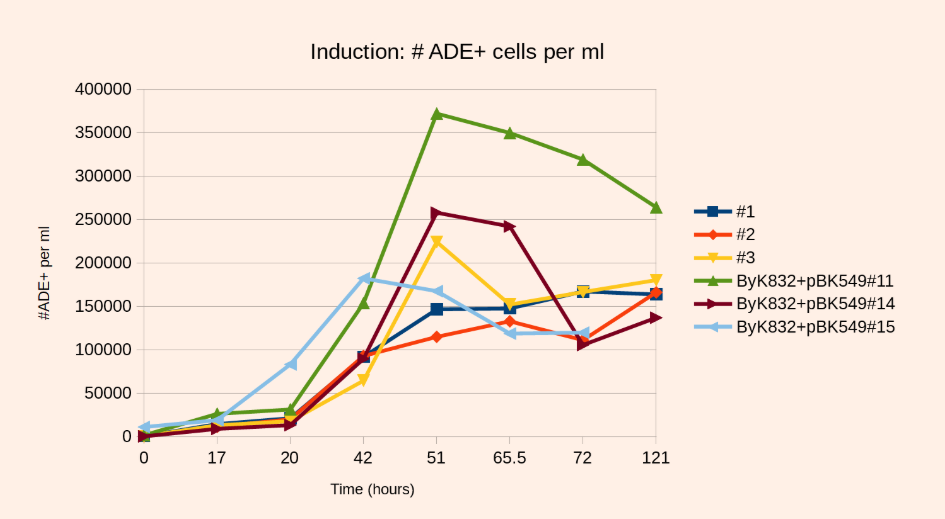
- Step 1: At the time of induction we plate 200ul from the induction culture at OD=0.2
We could not plate at T=0 from the induction culture, because we did not have enough plates.
- Step 2: Next day morning repeat Step 1 (17 hours)31072019 - We aborted this attempt because of a high background
- Step 3: 3 hours later repeat Step 1 (20 hours) - Step 4: Next morning repeat Step 1 (42 hours) - Step 5: Same day afternoon , repeat Step 1 (51 hours) - Step 6-8 : Every morning Step 1, for 5 days more.The OD should have reached 4-5 after 20h of induction.
Measure OD , next day after induction.
In order to know how well the transposition is going before reseeding:
- Spread 200ul directly on SD-ADE
- Spread 200ul of a 40000X dilution on SD and on SD-URA
- Score under a binocular after 30h (i.e. closer to the time of reseed)
- Expect around 0.05-0.1% of the cells to have become ADE+ . Incubate the plate for one more day for a final count.
- Pursue with the culture that gives the highest number of clones, starting from a minimum of background.
- 500-1000X more ADE+ clones at T=51 hours than at T=0 of inductions is in the ballpark
# Benoit Data
Strain Induction OD start OD stop Time # ADE+ cells/mL
at START# ADE+ cells/mL
at STOPstrain # 1 0.189 6.96 51h 4.05E+02 1.46E+05 strain # 2 0.198 7.17 51h 2.60E+02 1.15E+05 strain # 3 0.194 7.13 51h 2.85E+02 2.24E+05 strain # 4 0.196 7.11 51h 1.43E+02 3.72E+05 strain # 5 0.172 7.13 51h 2.25E+02 2.58E+05 strain # 6 0.199 7.27 51h 2.55E+02 1.82E+05 # Our data
Strain Induction OD start OD stop Time # ADE+ cells/mL
at START# ADE+ cells/mL
at STOPByk832T_3 (Leila) 0.098 51h Byk832T_1 (Enzo) 0.484 51h Byk832T_2 (Enzo) 0.510 51h Byk832T_3 (Enzo) 0.393 51h
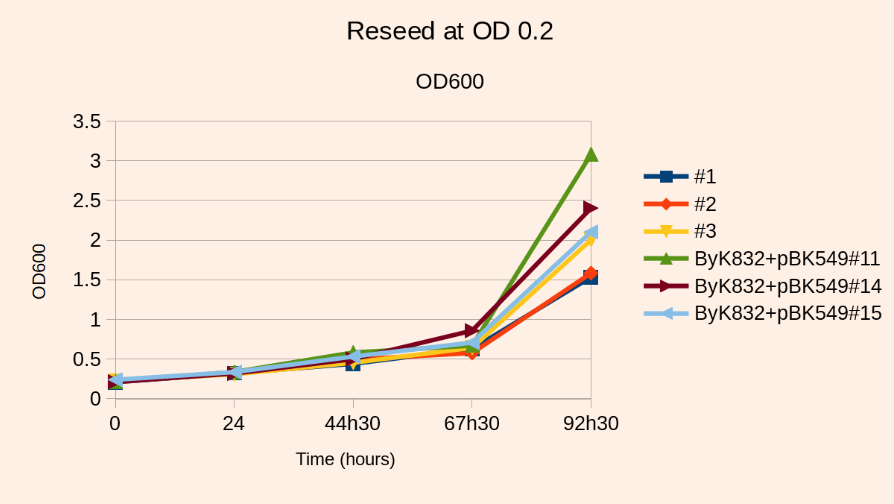
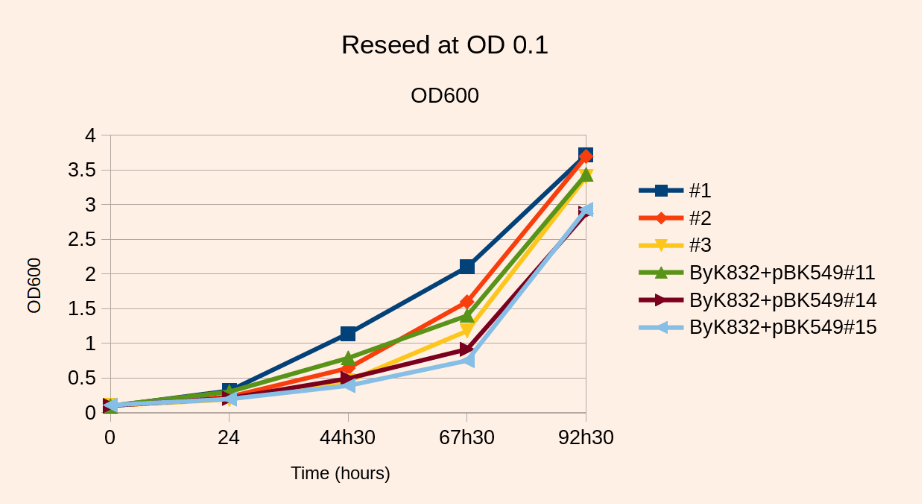
Reseed
- Determine the number of ADE+ clones after 50-52 hours of induction. If you have around 2.35E5 ADE+ cells per mL is GREAT.
- Inoculate 14E6 cells (14 million of cells) in SD-ADE 2% glucose at OD=0.2 in 2-4L
- Incubate at 30C, 140 rpm, harvest at OD~2 (80 hours). Note that the final OD will not exceed the value of 2.
- Fo reasons we ignore, the presence of ade- cells in the mix inhibits the growth of ade+ cells. This is why the culture will not grow above OD=2. This is also why you should not reinoculate at an OD above 0.2
- To estimate the growth of ADE+ cells: dilute the culture 2000X, spread 200 ul on SD-ADE – dilute 40000X, spread 200 ul on SD and SD-URA – Expect the number of ADE+ cells to have been multiplied by ~1000X.
Note1: If you don’t mind increasing the volume, you can reseed at a lower OD, and thus have less growth inhibition. But in any case you want to inoculate enough cells so as to keep a good complexity in your library.
- For example, a library containing 1.8E5 ADE+ cells/ml (OD=7.27, 0.15% ADE+ cells) after induction and reseeded at OD0.2 in 2L (1E7 independent transpositions) will reach an OD of 2 and a fold increase of 1250
– the same library reseeded at OD0.1 in 2L (5E6 independent transpositions) will reach OD3 for a fold increase of 3700
- Harvest cells and proceed to DNA extraction as in published protocol.
Because of the growth inhibition, only ~70% of the harvested cells are ADE+, meaning that during the sequencing reaction, 30% of the reads will map to the untransposed transposon in ADE2.