Title: 25062019-Experimental Card about the SATAY experiment
Contents
93. Title: 25062019-Experimental Card about the SATAY experiment#
93.1. What is the question?#
Main question:
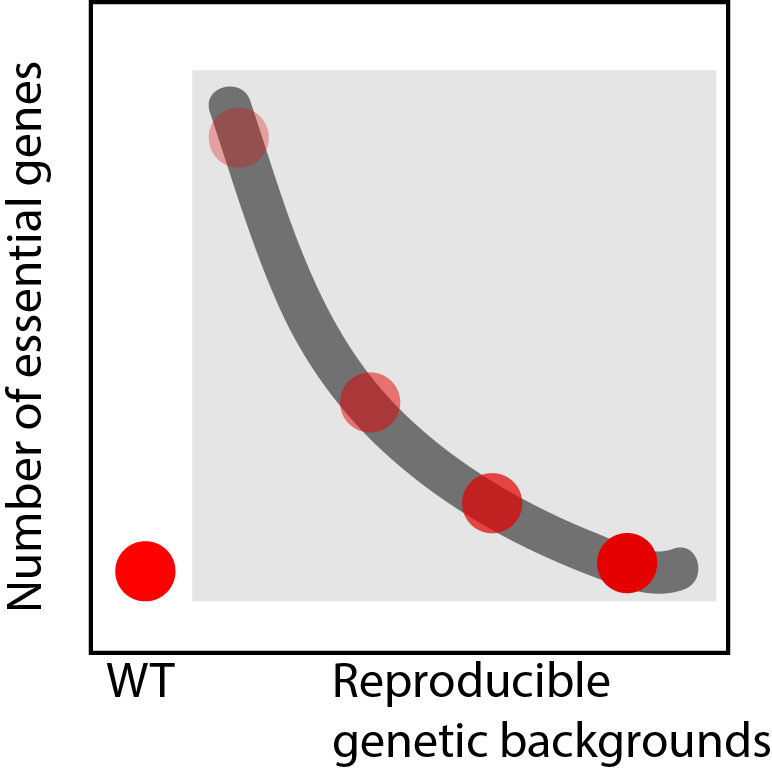
Are the reproducible genetic backgrounds acquired during evolutionary adaptation after crippling cell polarization showing a steadily change of essential genes towards the uncrippled cell essential gene profile?
Subquestions:
To what extent are the essential and non essential genes changing across genetic backgrounds, such that it can be used as observable for the global genetic interaction network “state”?
93.2. Why is it hard? / why it has not being solved before?#
The major hurdle in this question is that it implies existing high throughput techniques able to map the whole genome in different “states”.
Hence, how to map the yeast genome to search for essential and non essential genes in a systematic manner, is the central challenge for this. So far, what that have been used for measuring the essential genes of yeast genome are chemical mutagenesis methods, homologous recombination strategies, dCas9- based gene repression, among others. Most of them are tedious and incomplete for analysing the whole genome. Also, in some of these techniques, it is possible to get more than one mutation per mutagenesis step, which is not ideal in this case, because we lose control over the one to one gene interaction.
93.3. What would be my approach?#
The research question indicates that the essentiality property of genes can change depending the genetic background/environmental conditions present in the organism. It has been already studied that the essentiality property depends on the environment the cells experienced. However, the systematic measure of which genes are essential along different genetic backgrounds has not yet being tested due to the a lack of high throughput techniques to realize this.
Recently, a technique called Saturated transposition in yeast tackle this problem in a highly efficient way. The main principle is to saturate the yeast genome with independent insertions of the miniDs transposon. Deep-sequencing of the transposon-genome junctions allows to identify essential and non essential genomic loci. Since, the cells that happen to have a transposition event on an essential genomic loci wont grow, we wont have them on the sequencing library, hence those regions in the sequencing library will be devoid of transposon insertions.
Therefore, to tackle the question of finding whether for the evolutionary trajectory of bem1 deleted mutant, there is a recovery of essential genes towards the wild type ratio , I will applied the SATAY technique over meaningful intermediate steps (intermediate genotypes) in the evolutionary pathway, the parental mutant strain and wild type strain.
93.3.1. What will I measure?#
Transposition events in each of the reproducible genetic backgrounds, namely:
bem1Δ ura3- ade2-
bem1Δbem3Δ ura3- ade2-
bem1Δbem3Δnrp1Δ ura3- ade2-
bem1Δbem3Δnrp1Δbem2Δ ura3- ade2-
wild type ura3- ade2-
93.4. How a possible answer can look like?#
The evolution from a crippled cell towards a wild type phenotype cell requires/implies evolving towards the wild type profile of essential genes.