Title : PCR and sequencing checks for the new strains dbem3 and dbem3dnrp1
Contents
24. Title : PCR and sequencing checks for the new strains dbem3 and dbem3dnrp1#
24.1. Date#
25012021
24.2. Objective#
To check that the transformation were inserted in the right locations
24.3. Method#
Replate the strains in YPD plate (they were more than one month in the fridge)
Inoculation in 5mL YPD+6xade with NAT for dbem3 cells (colonies 10,11,12,13,14)and NAT+ HYGRO for 5 colonies from dbem3dnrp1 cells . (26012021)
Three tubes out of 5 from dbem3dnrp1 cells were dense the rest not, after 24h of incubation. They were not contaminated when looking through the microscope I only saw yeast cells.
Only one tube was dense for dbem3 cells ,and that was contaminated with bacteria. I looked the suspicious tube through the microscope.
Store in glycerol stocks from liquid culture
I stored the three dense tubes from dbem3dnrp1 cells (colony 2,10,11)
I did not store anything from dbem3 cells.
24.3.1. Colony PCR#
Use the colonies from the replate used to inoculated in liquid culture.
Design sequencing primers
spy6/7 from Els(bem3)(oLL401/402)
spy1/2 from Els(nrp1)
1uL template (one colony in 50mL)
I wrongly used 1uM from oLL401/402 instead of 10uM because I diluted 10x from a 10uM stock ….
I used both set of primers in all the templates.
27012021: repeat the PCR with the right primers concentration and the same colonies I have kept in MiliQ in eppis.
use 5ul of template
Restreak more colonies from the original selection plate from the transformation for dbem3 on YPD+NAT plate.
I restreak like 16 colonies more and surprisingly they showed very miserably growth after a weekend growth period.
PCR on the three more grown colonies with SPY1/2 and oLL401/402 primer sets, and the same protocol as before. I used 5ul template.
Try again same PCR with 1ul template
Incubate in liquid media YPD+6x+NAT single colonies from the streak 1, 2 and 3 from dbem3 colonies. Also incubated the colony 1 mixed in MiliQ used for colony PCR in YPD+6x+NAT media to compare the consistency from the colony taken from the streak 1.
The only grown culture was from colony 1 from the plate. The one from the eppi did not grow (?)
Colony PCR with the culture with colony 1.
550ul of 2x washed liquid culture in MiliQ and resuspended in 50uL MiliQ. I took 1uL template for PCR.
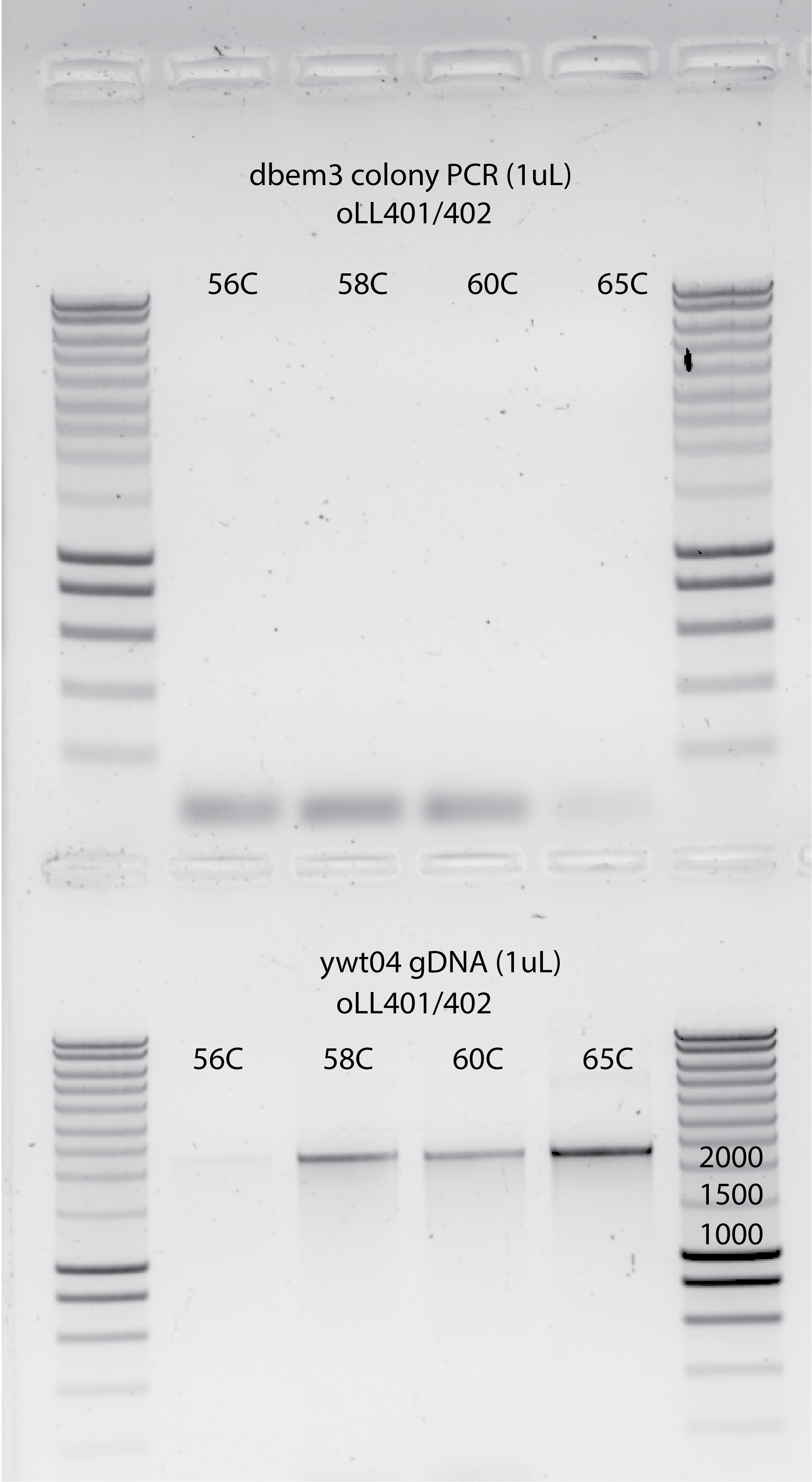
PCR protocol: annealing temperature gradient to find the optimal conditions for this PCR. (56C,58C,60C,65C) and also increasing to 3mins the step with 98C which is the one who breaks the cell wall . In the previous experiments was set to 30secs, so maybe it is not sufficient to get enough template.
I also used 1uL gDNA from ywt04 as a positive control with oLL401/402 primers.
24.4. Results#
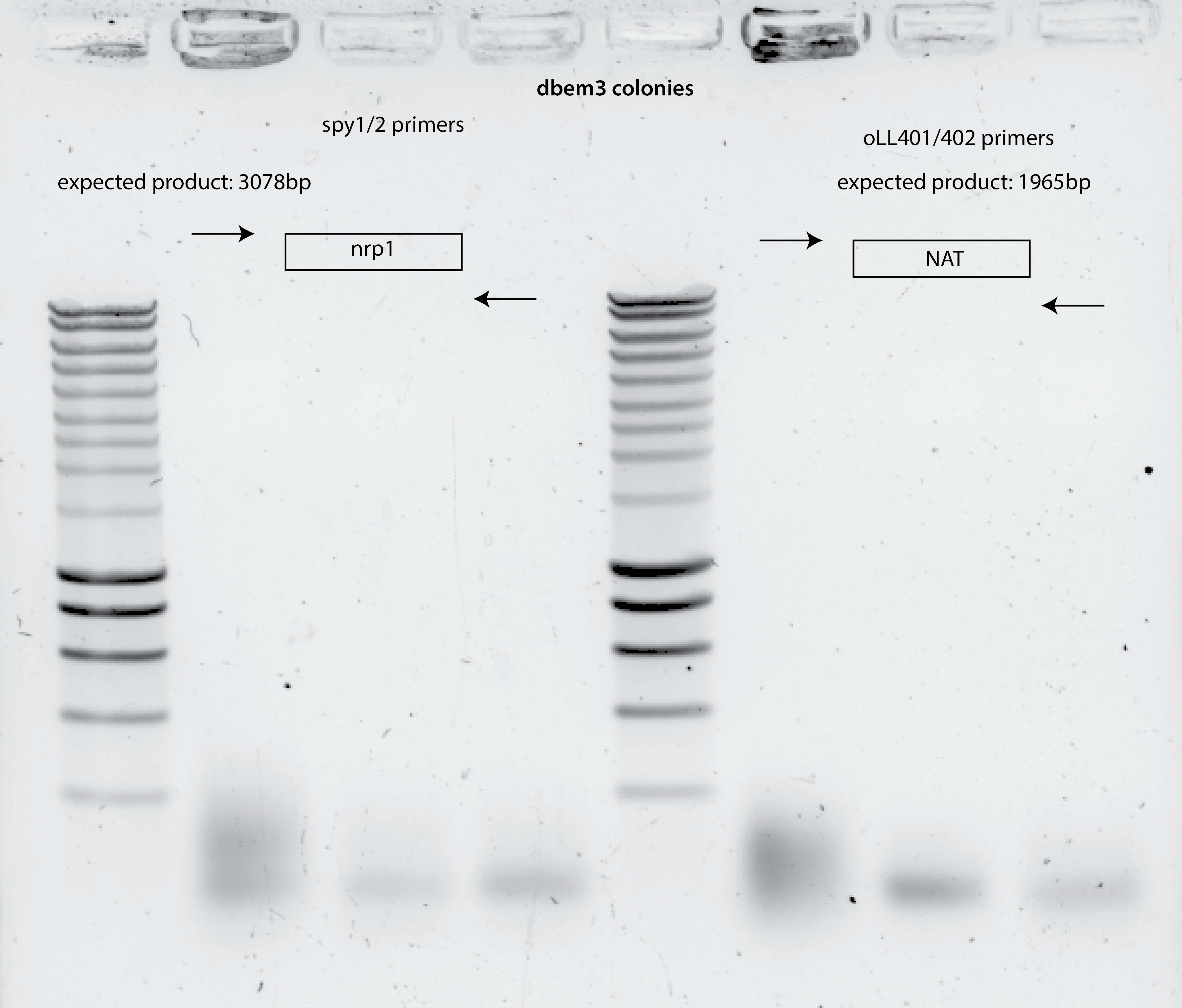
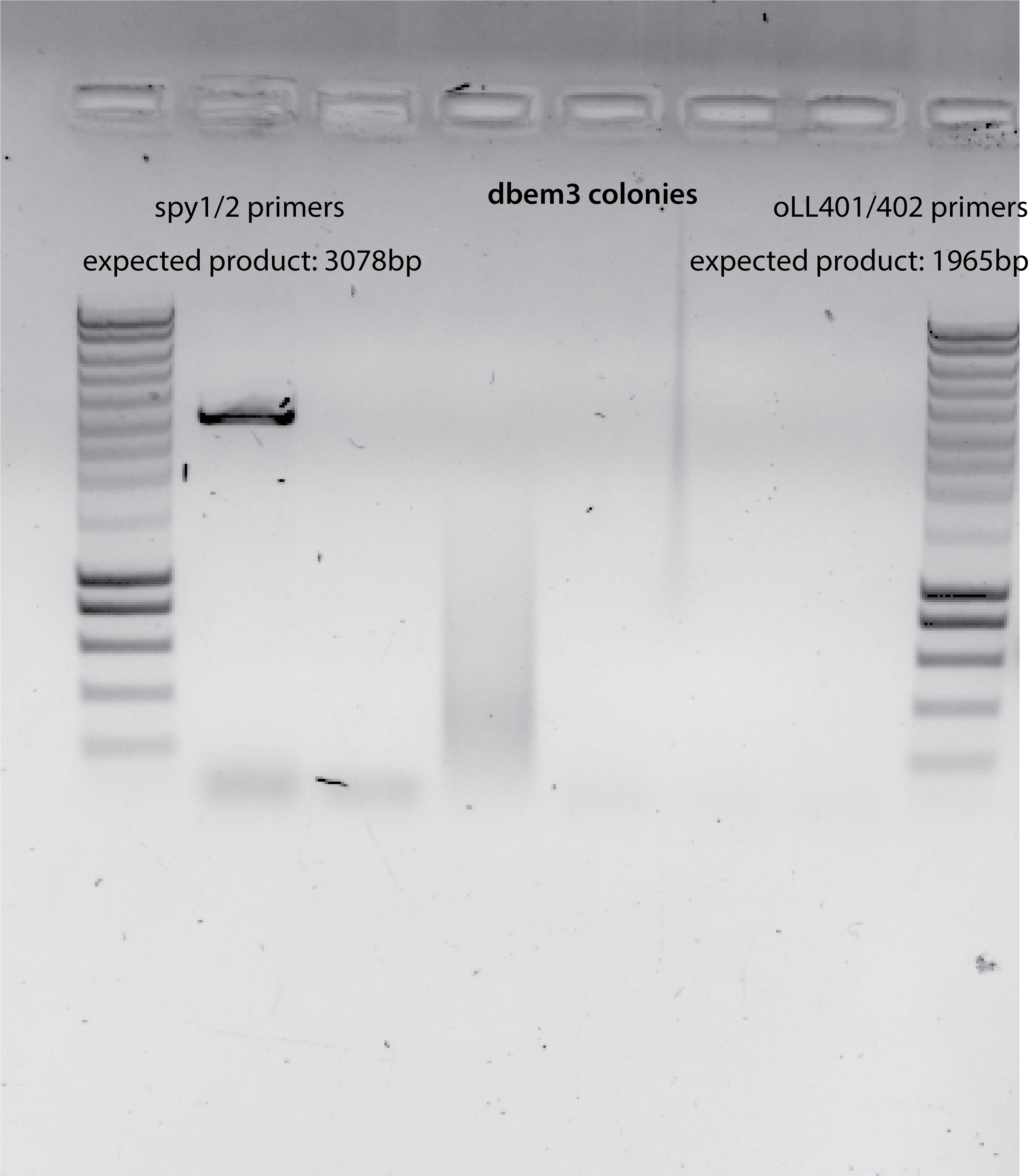
No product from PCR

Confirming three clones from dbem3dnrp1 strains (colonies 2,10 and 11). The same clones that grew on liquid media YPD+NAT+HYGRO :)
Glycerol stock storage: ylic138 a,b,c (3 biological replicates)
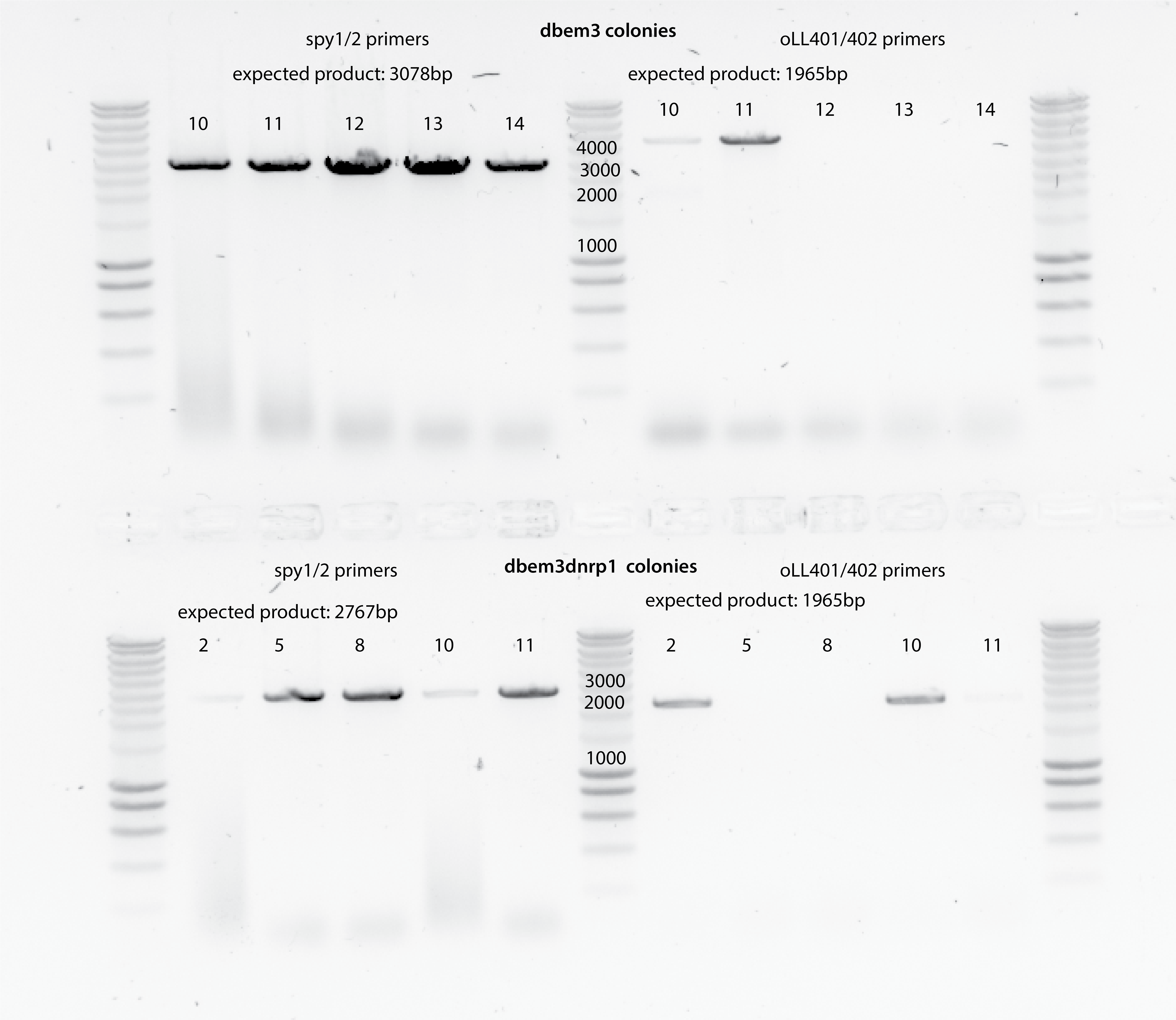
All checked bem3d colonies are not correct because the band shown for the oLL401/042 primers indicates that BEM3 is present in them.
NO PCR PRODUCT for dbem3 taken colonies , 5ul template.
NO PCR PRODUCT for dbem3 taken colonies , 1ul template.
NO PCR PRODUCT for dbem3 colonies. Correct positive control
24.5. Next Steps#
Sequencing of correct mutants on bem3::NAT transformation.
Look if colony 10 and 11 from dbem3 transformation are actually the big colonies or the small, and also for the rest that did not show any band to correlate fitness with colony size and correct transformation.
colony 10 and 11 seem to be actually the normal size colonies (not small)
Do a PCR for transformation (again on the WT background to get bem3d) from the dbem3dnrp1 strain around bem3::NAT with like 1000bp of flanking regions downstream and upstream the NAT . The longer the flanking regions the more likely is to insert in the right location in the genome. FIRST CHECK WITH gDNA FROM dBEM3 STORED STOCKS IF IT DOES NOT WORK .
design of the right primers (olic54 and olic55)
