Title : Contruction dbem1dbem3+pGal from ylic139 -> ylic140
Contents
17. Title : Contruction dbem1dbem3+pGal from ylic139 -> ylic140#
17.1. Date#
30062021-15072021
17.2. Objective#
To get a clean dbem1dbem3+pgal:CDC42 because ywkd073I and II is growing miserably.
17.3. Method#
Transformation protocol on ylic139: ywkd071+bem1:KAnMX
Use ywt04gDNA with oll401/402 primers, bem3::NAT to get the PCR DNA for transformation.
1ul ywt04 template
Leila_60 PCR protocol
8 PCR to merge and have more DNA
make 10 SC-URA(4x)+ 2%Raff+2% Gal + G418+NAT as selection plates for the transformation.
Incubate 10ml ylic139 in 4x CSM+2% Raff +2% Gal + G418 (20ul 500X) (11:00)
PCR purification , 4 eppis , elution 20ul for each eppi. (I used 200ul for binding buffer since the product is bigger than 2000kb)
Note : I should have used primers olic51/52 to build the DNA for transformation because primers olic401/402 are more used to check the location of the insertion after transformation.
same PCR with olic51/52 for transformation
the gel did not show any band …. I dont get why…
repeat()
17.4. Results#
Expected size for the PCR (though the gel was not great)
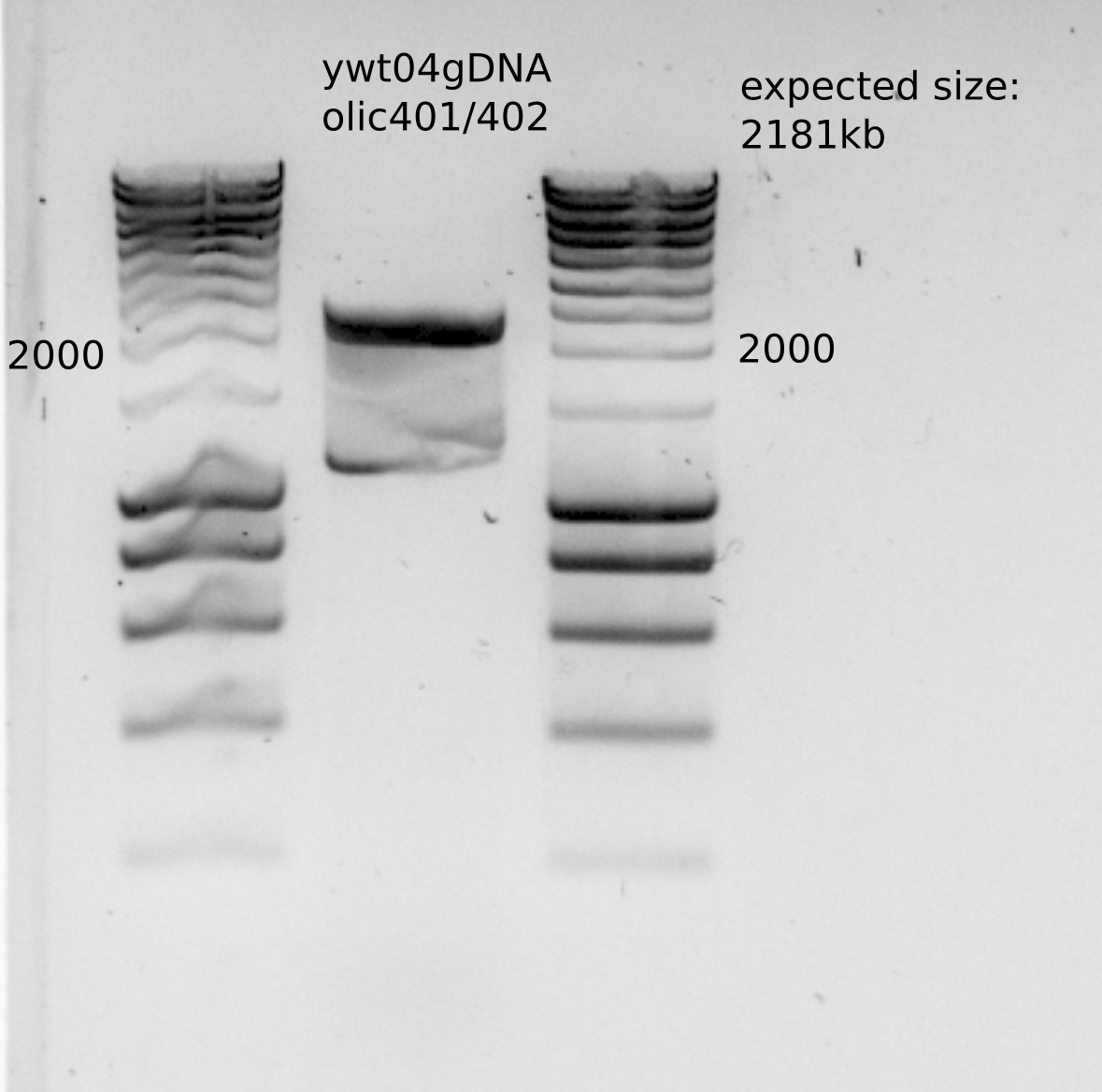
Purified PCR concentration: 87 ng/ul , having around 75ul , which gives as a total amount approximately : 6.5ug
For Transformation I should around 2ug , so around 25ul .
Expected size for the PCR with olic51/52
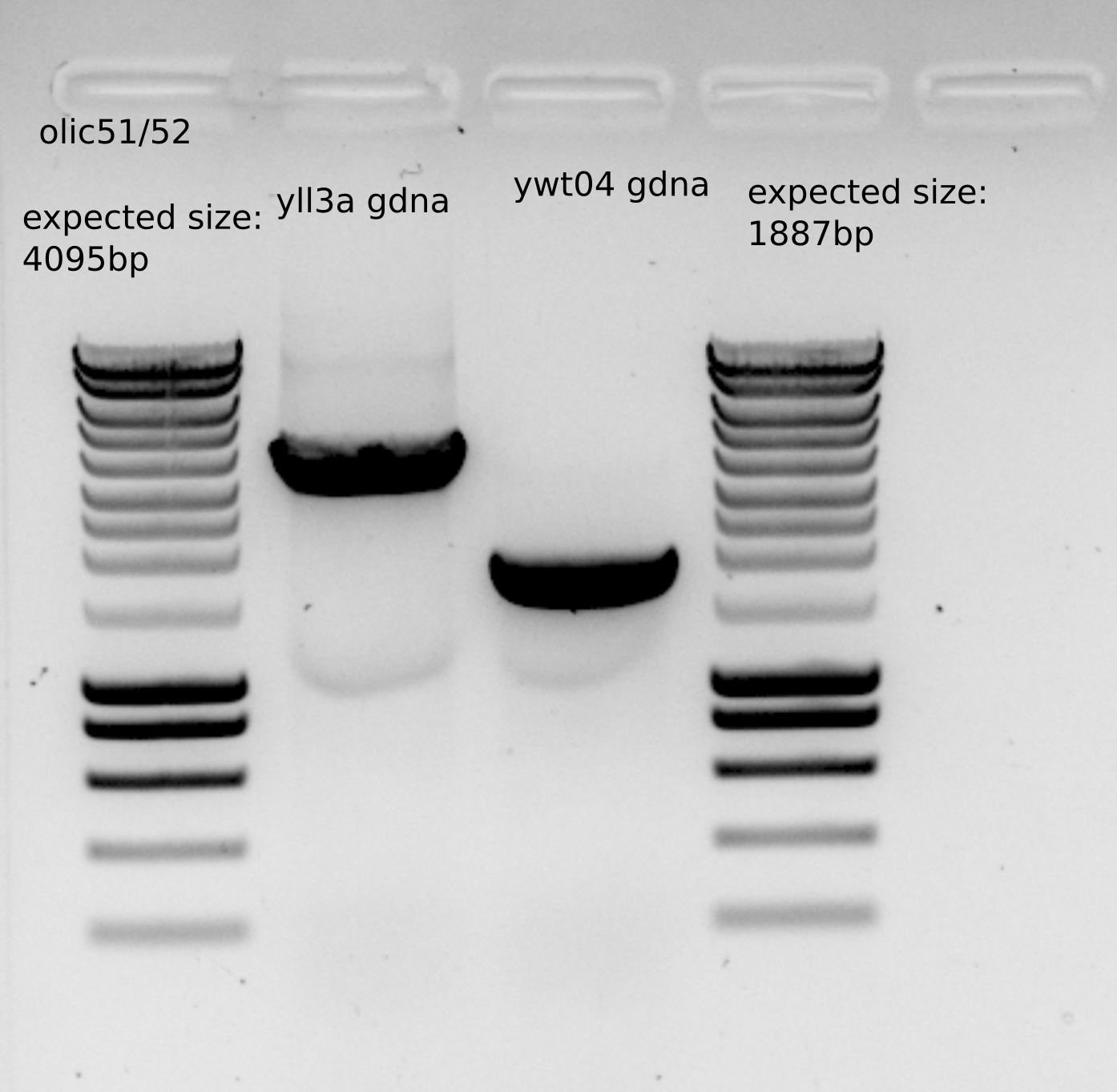
Purified PCR concentration: 90 ng/ul , having around 40ul , which gives as a total amount approximately : 3.6ug
For Transformation I should around 2ug , so around 22ul .
17.4.1. Incubation results#
Not growth after overnight liquid incubation by ylic139.
Change of strategy: Plate ylic139 in CSM plates with Gal and G418 and take a single colony to inoculate in liquid culture. (01072021)
I used already made CSM+0.1%Gal that I added 1.5ml of 20% gal . Once they get dry I will add 30ul 500x G418 and streak the cells from glycerol stock.
There were cells on the plate after growth during weekend, so I took two single colonies, from ylic139a and ylic139b and inoculate in 4xCSM-NF+2% Raff+2% Gal+G418 500x(20ul) at 9:30.
17.4.2. Transformation#
OD measurement on 06072021
Time |
Strain |
OD- 2x diluted |
real OD |
|---|---|---|---|
11:00/06072021 |
ylic139_a |
0.275 |
0.55 |
ylic139_b |
0.137 |
0.27 |
|
15:00/06072021 |
ylic139_a |
0.279 |
0.56 |
ylic139_b |
0.222 |
0.44 |
|
08:45/07072021 |
ylic139_a |
0.3 |
0.6 |
ylic139_b |
1.127 |
2.24 |
Transformation with PCR (ywt04/olic51/52) on 07072021
25ul of DNA from purified PCR -> 25ul*90ng/ul=2.25ug
Recovery step , from 10:30->12:30 in 4x CSM+2% Raff +2% Gal
Plating:
selection plates: SC-URA(4x)+ 2%Raff+2% Gal + G418+NAT
After the recovery I centrifuged the samples and resuspend them in 100ul MiliQ .
I plated the transformed cells 50ul from this tube , 0X dilution.
I added 950 ul MiliQ to the rest and plate 50ul in one plate and 200ul in other one.
Plate 200ul in CSM+0.1% Gal to check viability.
Plate 50ul of 0x non transformed cells in selection plates.
Plate 200ul of 10x non transformed cells in CSM+0.1% Gal to check for viability.
Transformation plates
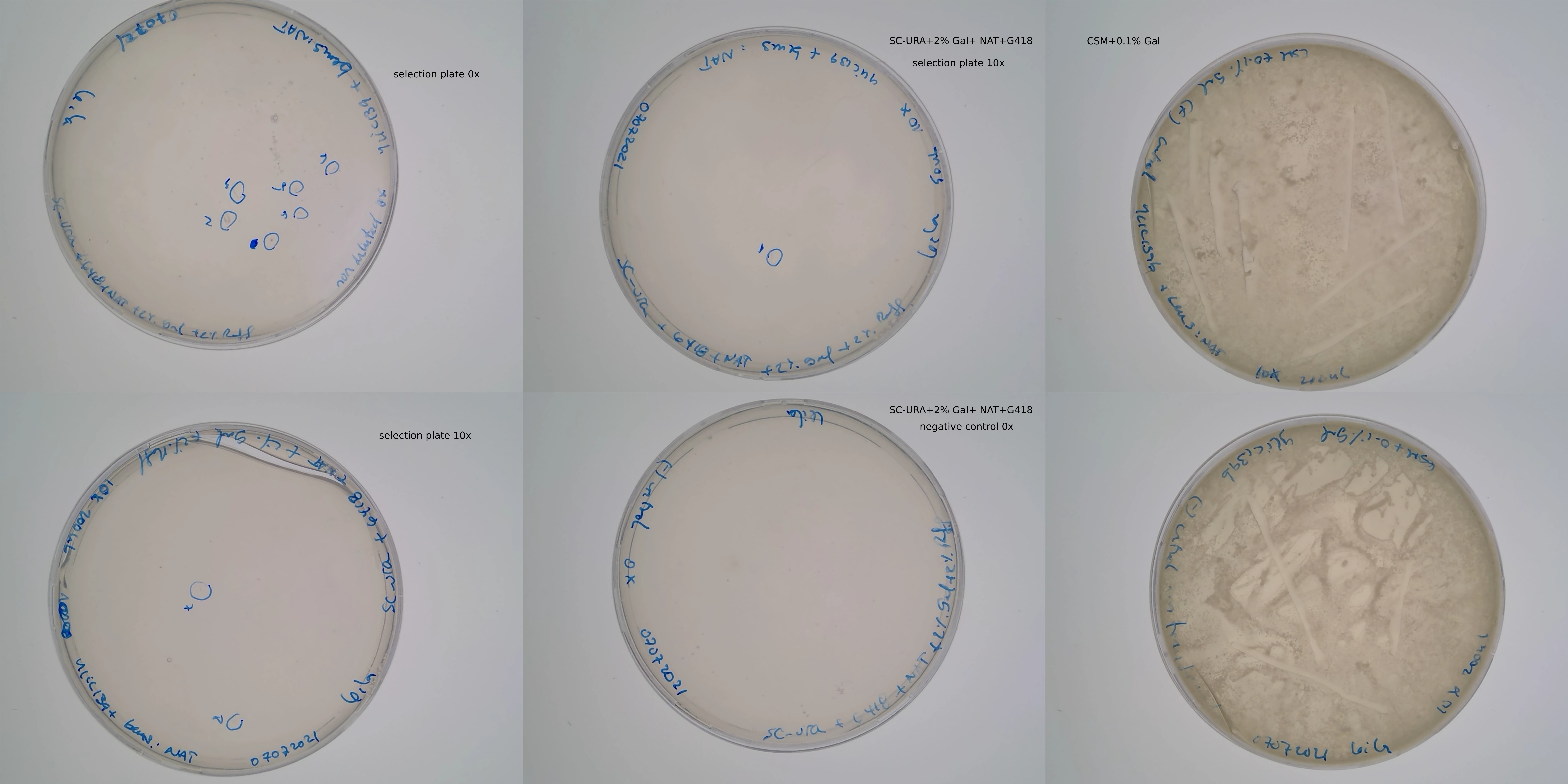
Very few colonies , it seems it was a hard transformation.
Colony PCR
8 colonies, colony 2 and 3 were big , the rest small.
Transfer colonies/ streak into CSM+0.1% Gal + NAT
5ul MiliQ suspension, 10ul MiliQ for the big colonies
1ul used for PCR (LEILA_60)
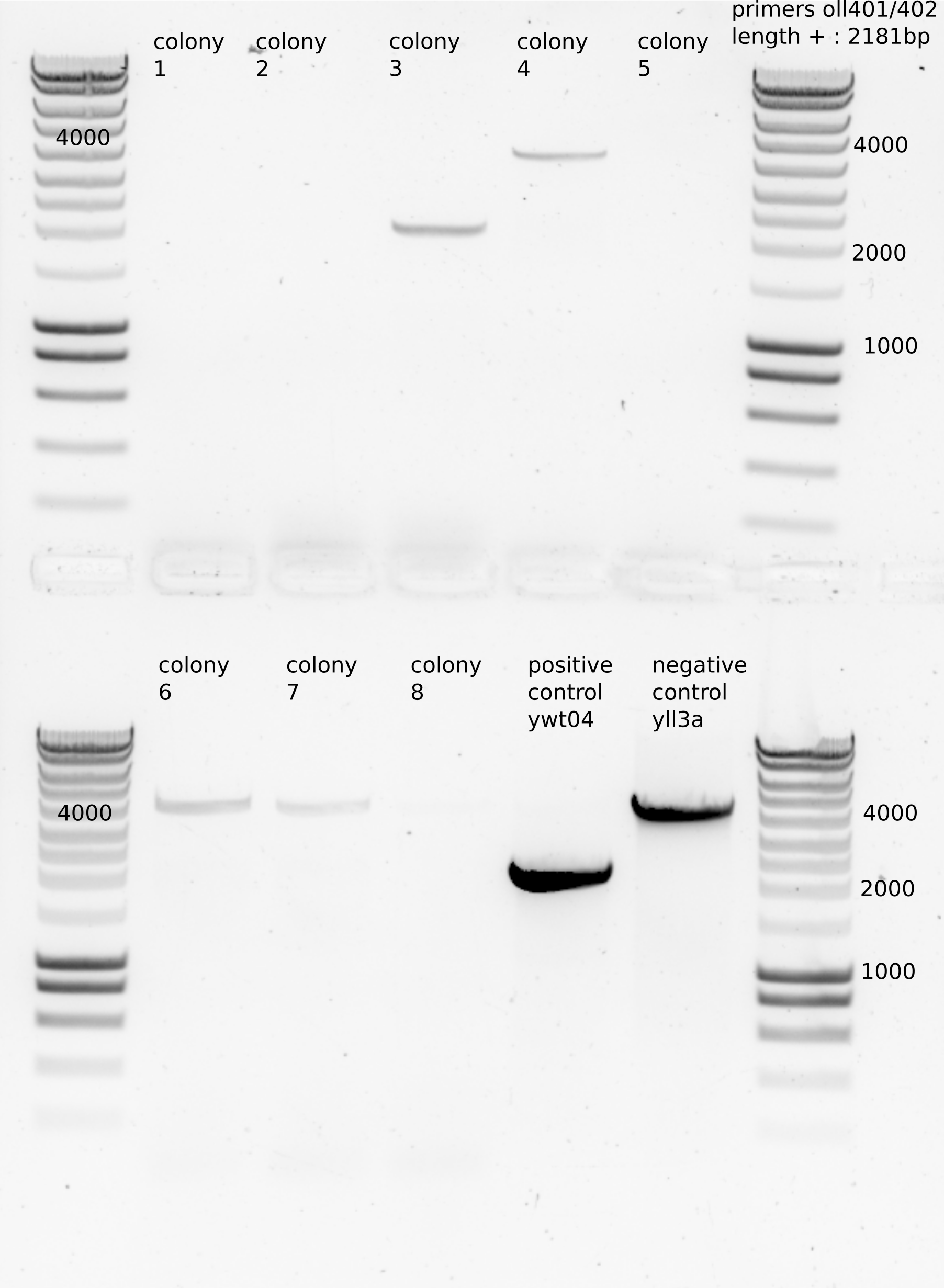
colony 3 seems the only right , which was a big colony. See the growth in CSM+0.1% Gal +NAT to store it in glycerol stock.
Re-streak and liquid incubation
restreak of the colonies for PCR in CSM+0.1% Gal +NAT
Inoculate colony 2 and colony 3 (the ones that grew and big colonies) in CSM+2% Gal+G418+NAT (5mL) for glycerol stocks.
17.4.3. PCR with gDNA#
DNA extraction of ylic140a (colony 2) and ylic140b (colony 3)
Inoculation in 10ml of 4X-NF-CSM+2% Gal+NAT+G418
Extraction of 4 eppies with pellet from 2ml of culture.
Elution of 20ul per eppi.
PCR
Check with oLL401/402 primers
Check with olic56/57
Include ylic139 as a negative control for the presence of BEM3::NAT (expected length around 4kB) and as a positive control for BEM1::KanMX (around 3kb)
5ul loaded of gDNA
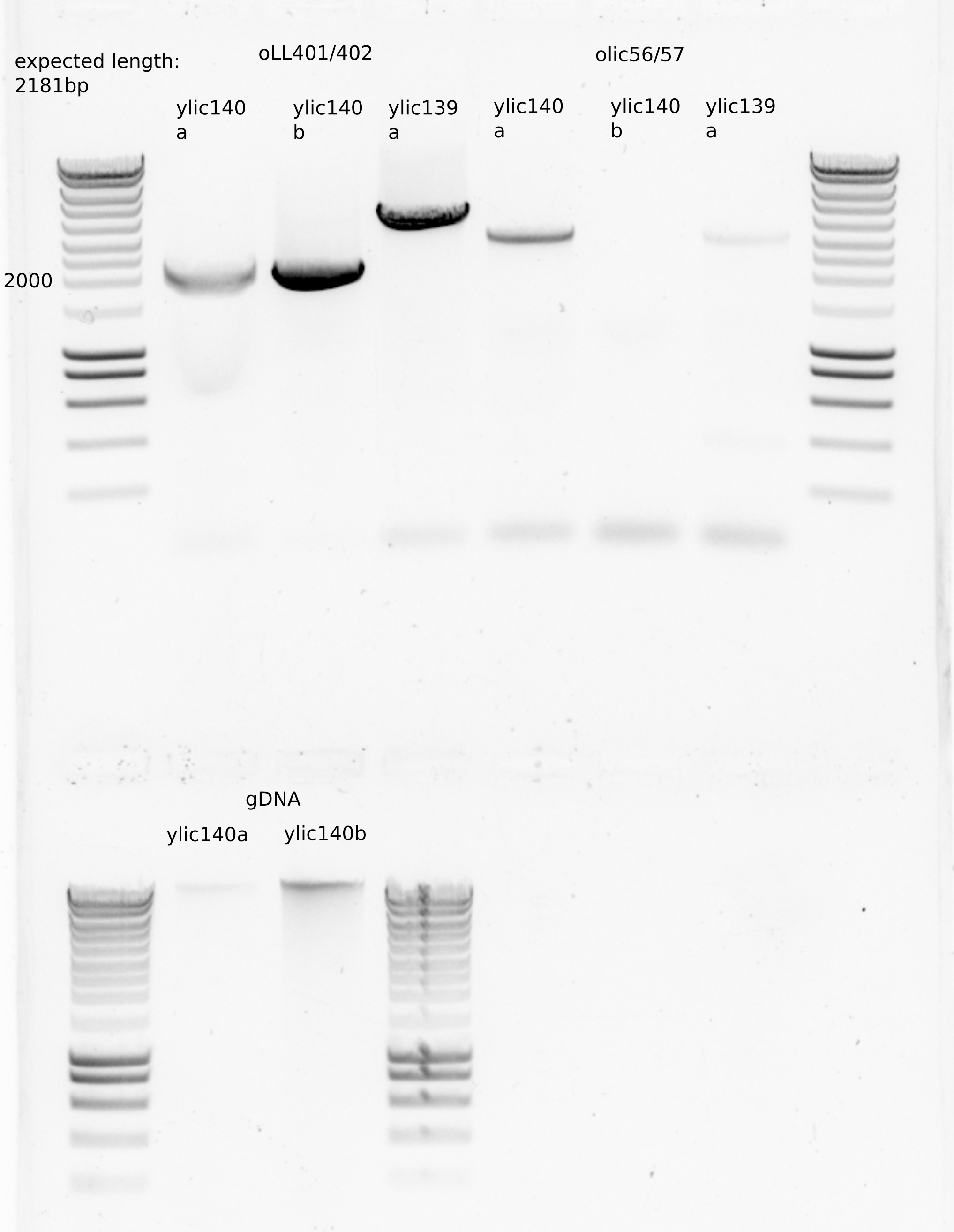
17.5. Conclusion#
The dna is ready for transformation
At least one colony is correct .
strain name : ylic140 MAT \(\alpha\) can1-100 leu2-3,112 his3-11,15 BUD4 from S288C bem1::KanMX bem3::NAT CDC42::URA3-GAL1pr-CDC42,CAN1::MFAprHIS
two biological replicates , a (colony 2 check again by PCR) and b (colony 3 checked by colony PCR)
