Title : gDNA extraction from yWT04a
Contents
28. Title : gDNA extraction from yWT04a#
28.1. Date#
27112020
28.2. Objective#
To have the DNA template bem3\(\Delta\)::NAT to make the PCR for transformation on ylic133 and ylic136.
28.3. Method#
Genomic extraction Kit with the Roboclon KIT.
Pellet from 2mL of culture in each eppi. I had 6 eppies.
30uL of elution.
concentration from the Nanodrop: 36ng/uL , much higher than before because of the activation of the column with Buffer BG that previously I was overlooking.
28.3.1. PCR#
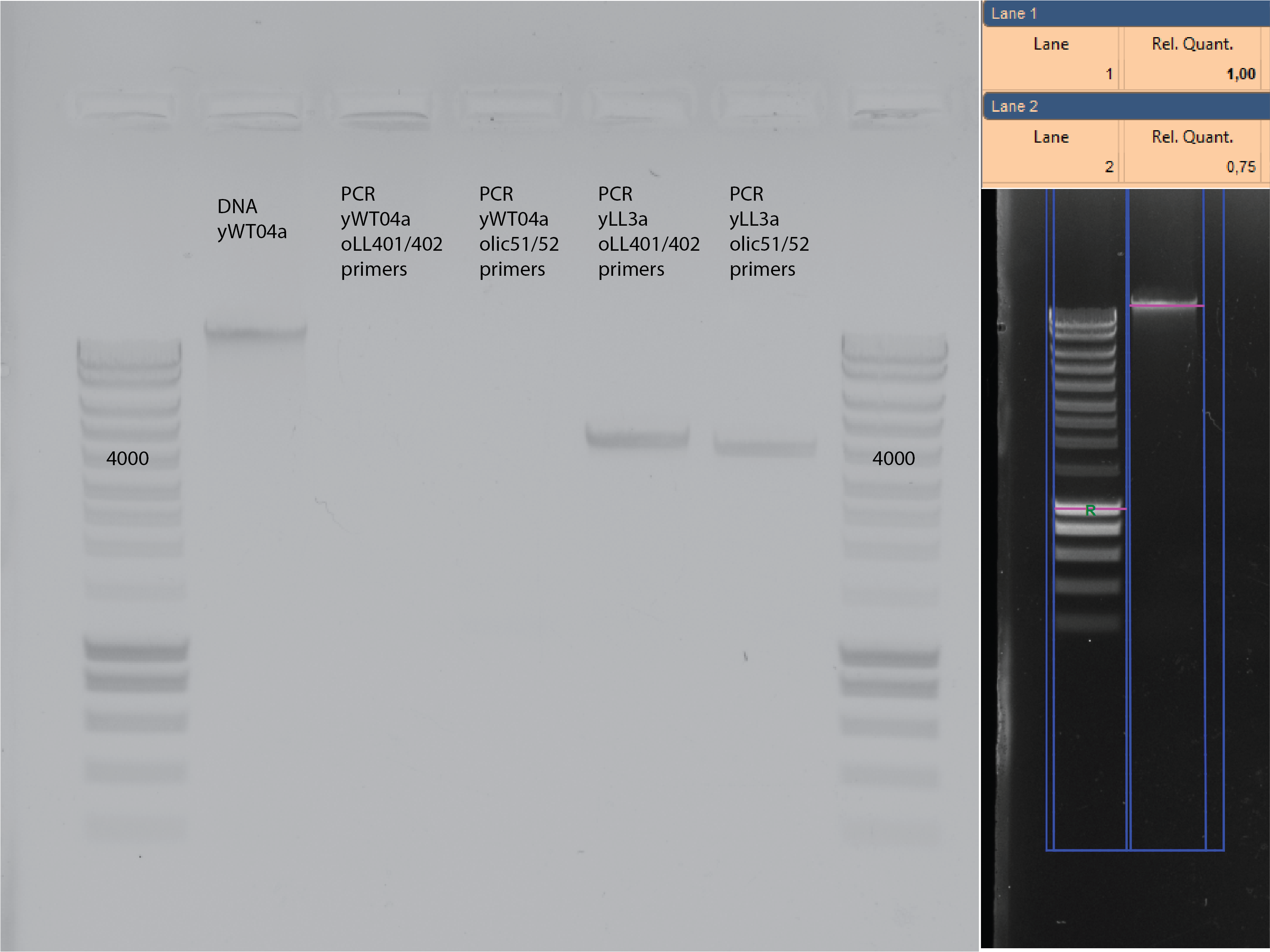
1ul of the DNA from ywt04a and yll3a reference
Primers 51/52 and oLL401/402
60C annealing temperature and 2mins 72C extension.
Gel 120V, 30 mins. 5ul loaded
28.4. Results#
28.4.1. Next steps#
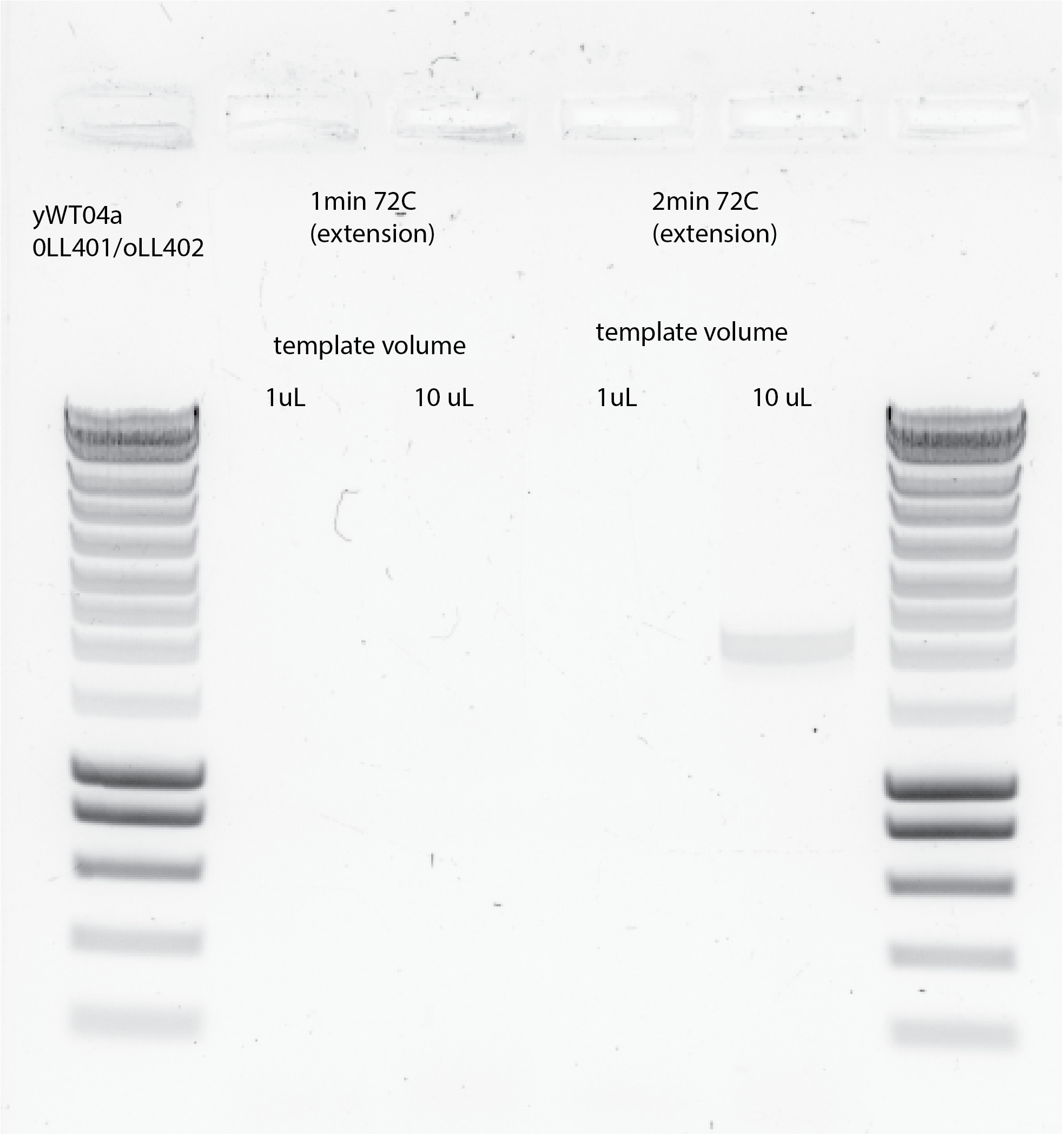
Two PCRs protocol with 1ul and 10ul ywt04a template with oLL401/402 primers using Wessel PCR protocol with 1min in the annealing temperature, instead of 30secs which is how he originally used (no reason). The differences between the two protocols is the extension time, in one is 1min and the other one 2mins.
3mins in 98C
30X(30s in 98C, 1min in 65C, 1min in 72C)
12 mins in 72C
4C on hold
 {width=50%}
{width=50%}
Repeating the PCR with 34X cycles , 10uL template and 2mins 72C extension. (8X) for transformation. (I used the pCR machine from downstairs)
It did not give any product (super weird!)
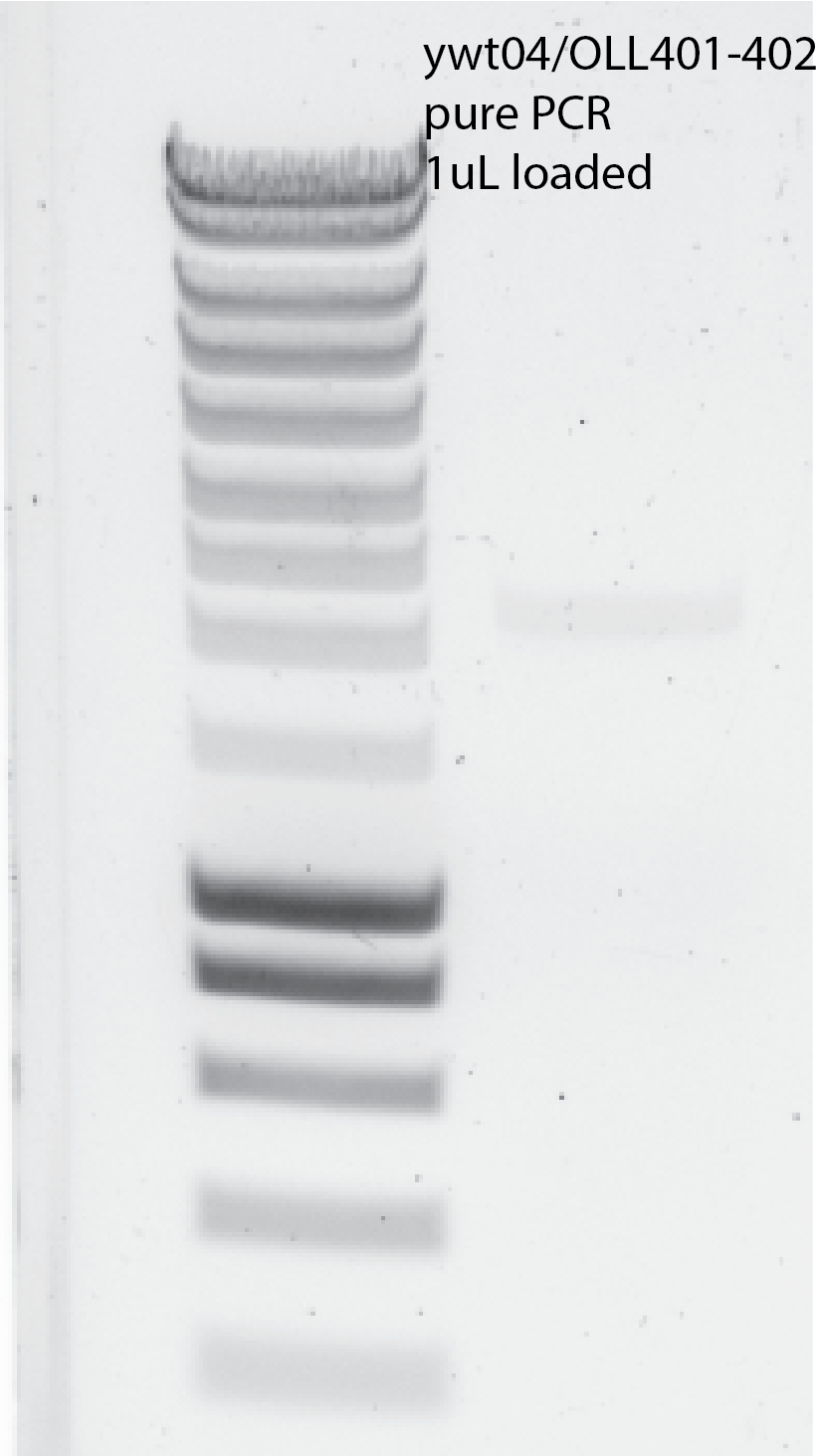
I pulled everything together with the previous PCR and purify it (10uL elution) and I got still very low yield ~8ng/uL.
 {width=50%}
{width=50%}Repeat the PCR with the same PCR machine as the one that worked.
I reduced the time in the annealing temperature from 1min to 30secs. This actually does not make any influence on that. (It did not work - I did not get any bands)
Repeat the genomic extraction (I ran out of DNA)
I cultured 2 tubes of 10mL to do completely.
 {#fig:DNA-extraction-good}
{#fig:DNA-extraction-good}
Nanodrop |
On Gel (BioRad) |
|
|---|---|---|
DNA-ywt04a |
60ng/uL |
180ng/uL |
very high yield of DNA !! :)
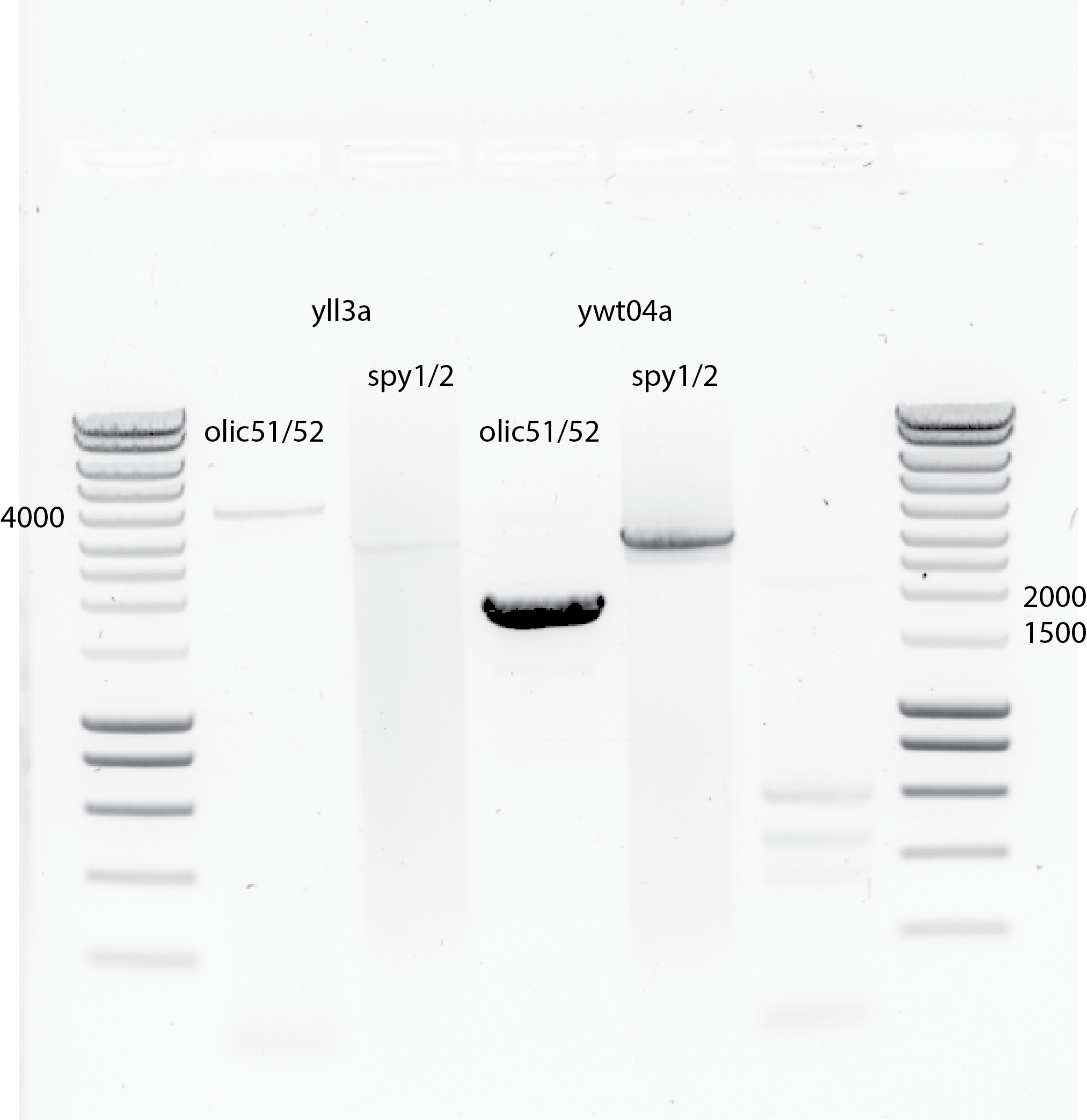
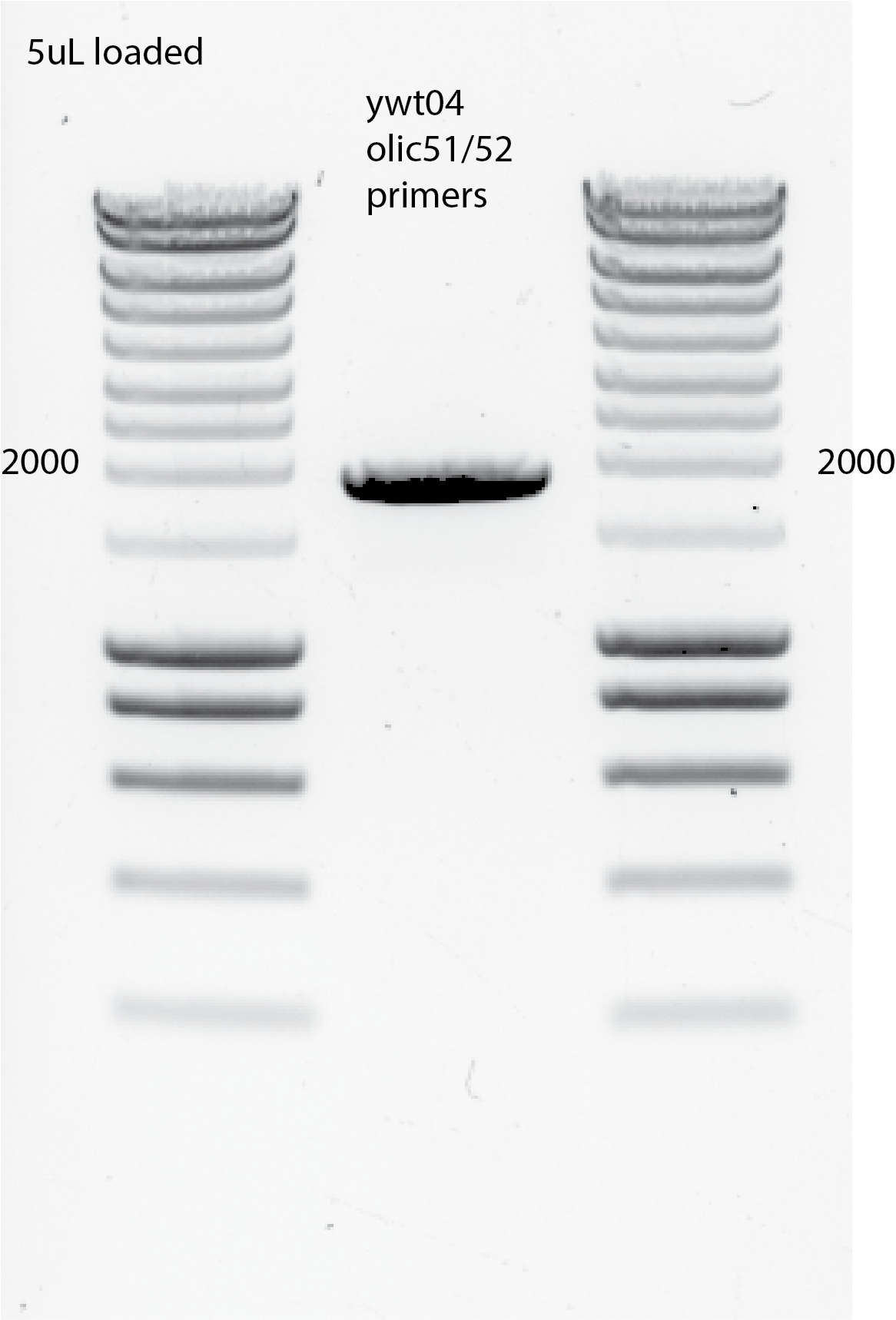
PCR using 1uL yll3a , 1uL ywt04a and olic51/52 primers and spy1/spy2 primers from Els as a positive control.
I changed the annealing temperature to 60C , 30 secs
Do 8 PCRs with 1ul ywt04a template and with olic51/52 primers for transformation.
PCR protocol: 60C annealing T , 30 secs ; 72C extension 2 mins; 98C denature DNA 3 mins
 {width=50%}
{width=50%}
28.5. Conclusion#
Using ywt04 as template did work with my primers olic51/52.
The last DNA extraction was super successful , and what I did differently was:
Using 20ml of culture
Pull the pellet of 2ml (I had 10 eppies in total)
Mix by inversion several times in the step of 55C incubation with proteinaseK for 30mins. Before I have never mix the tubes but this time I did , and I got very clean DNA as you see in [@fig:DNA-extraction-good]
The PCR are extremely sensitive sometimes with the conditions , in this case I have found that the most optimal conditions are:
98C 3mins
do 34x cycles:
98C 30secs
60C 30secs
72C 2min
end cycle
72C 10mins
12C on hold
The concentration by the Nanodrop of the purification of the 9 PCRs , using 10ul of elution distributed in 6 tubes , so in total around 60uL of DNA was: 260ng/uL :) . So I have around 13ug of DNA for transformation.
