Title : do yeast cells show multinuclei upon CDC42 depletion? (experiment)
Contents
7. Title : do yeast cells show multinuclei upon CDC42 depletion? (experiment)#
7.1. Date#
11102022 (Tuesday)- 17102022 (Monday)
7.2. Objective#
Proof that CDC42 depletion causes multinuclei in yeast cells.
7.3. Method#
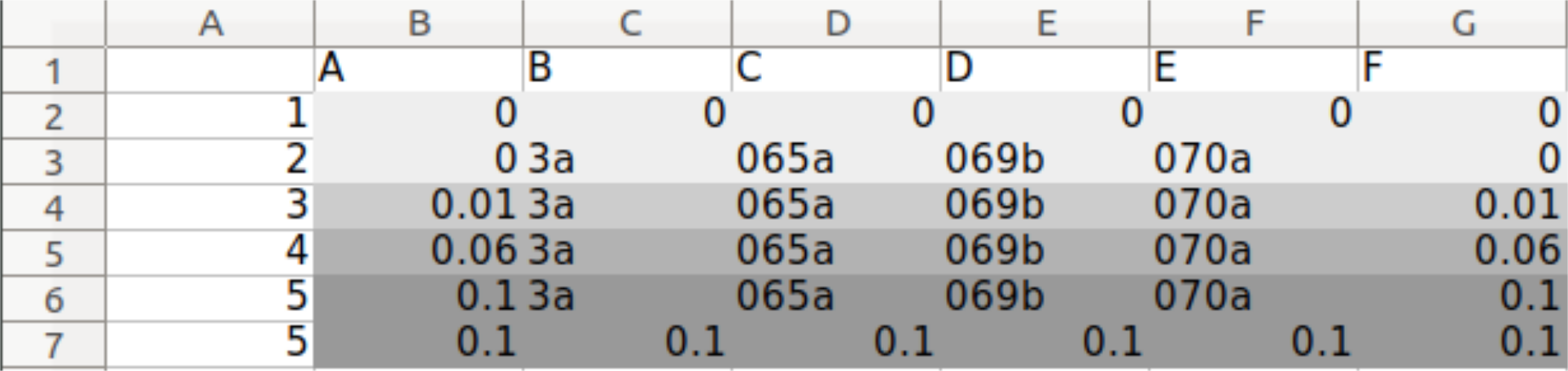
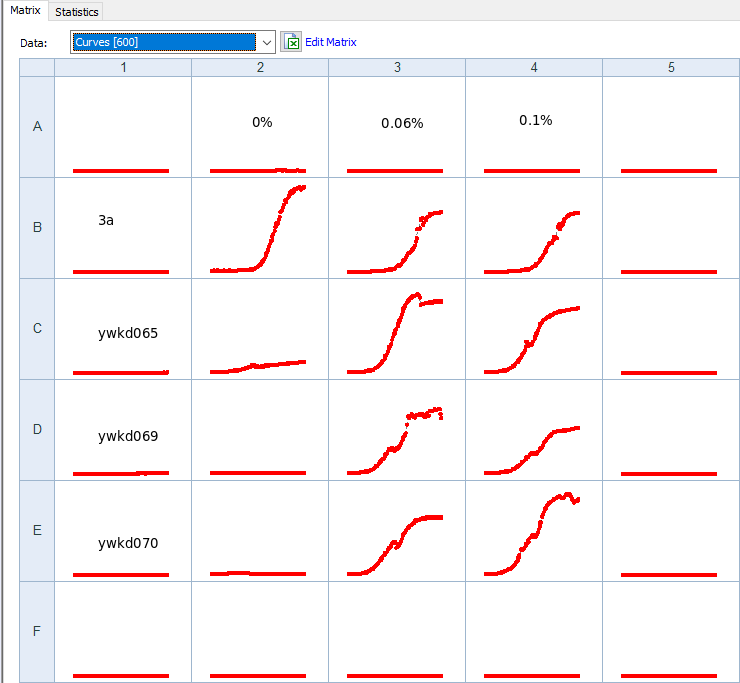
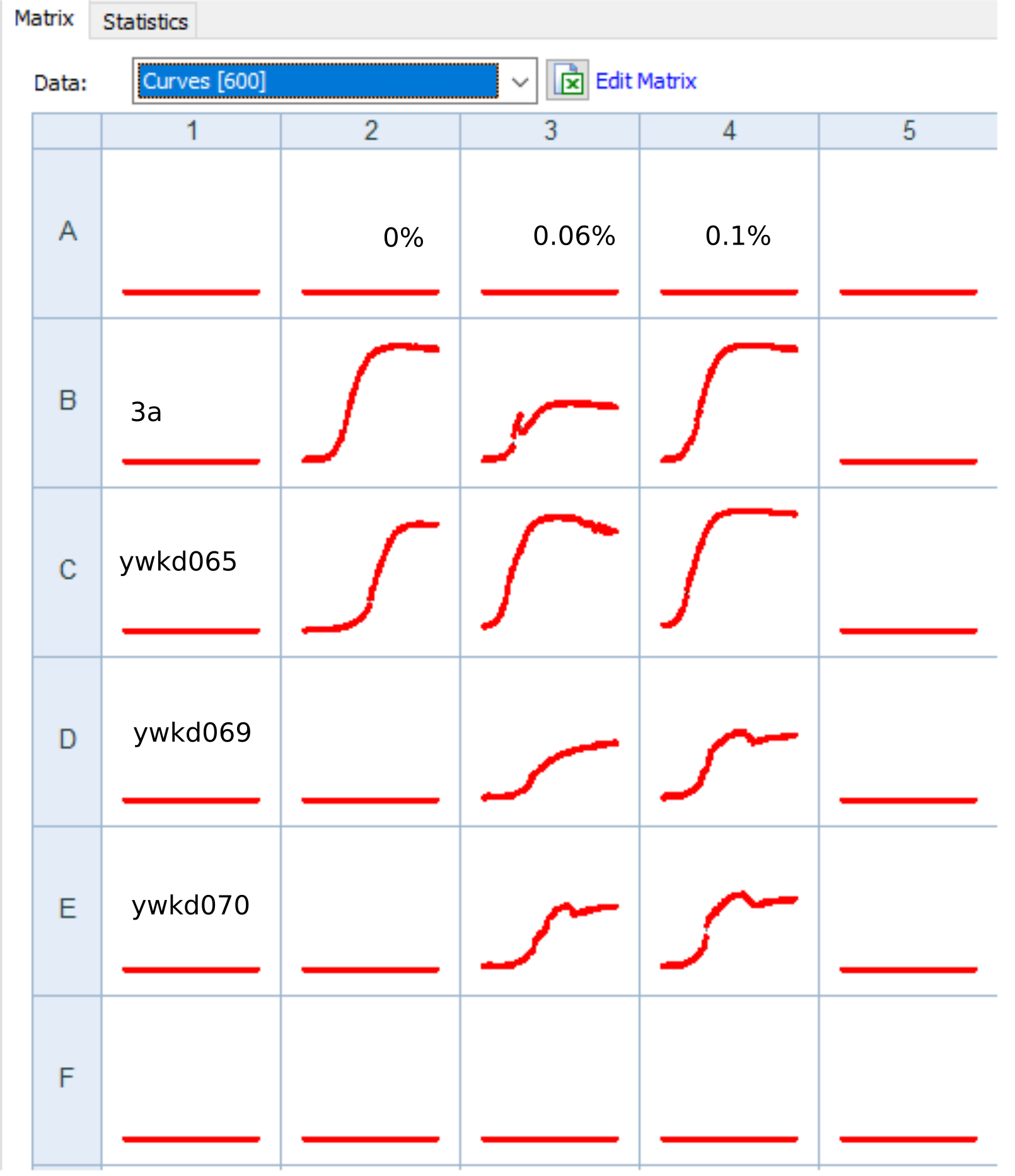
Perform population growth measurements in 0,0.06,0.1 % galactose for the following strains: WT+sfGFP+pGal-CDC42(ywkd065a),dbem1+sfGFP+pGal-CDC42(ywkd069b),dbem1dbem3+sfGFP+pGal-CDC42(ywkd070a),WT(yll3a Control).
In saturation, stain the cells with DAPI (follow this protocol)and take some fluorescence images.The images have to be taken with freash staining , meaning with less than 30 mins after staining, otherwise the cells will be fully broght because there is no washing step of the DAPI.
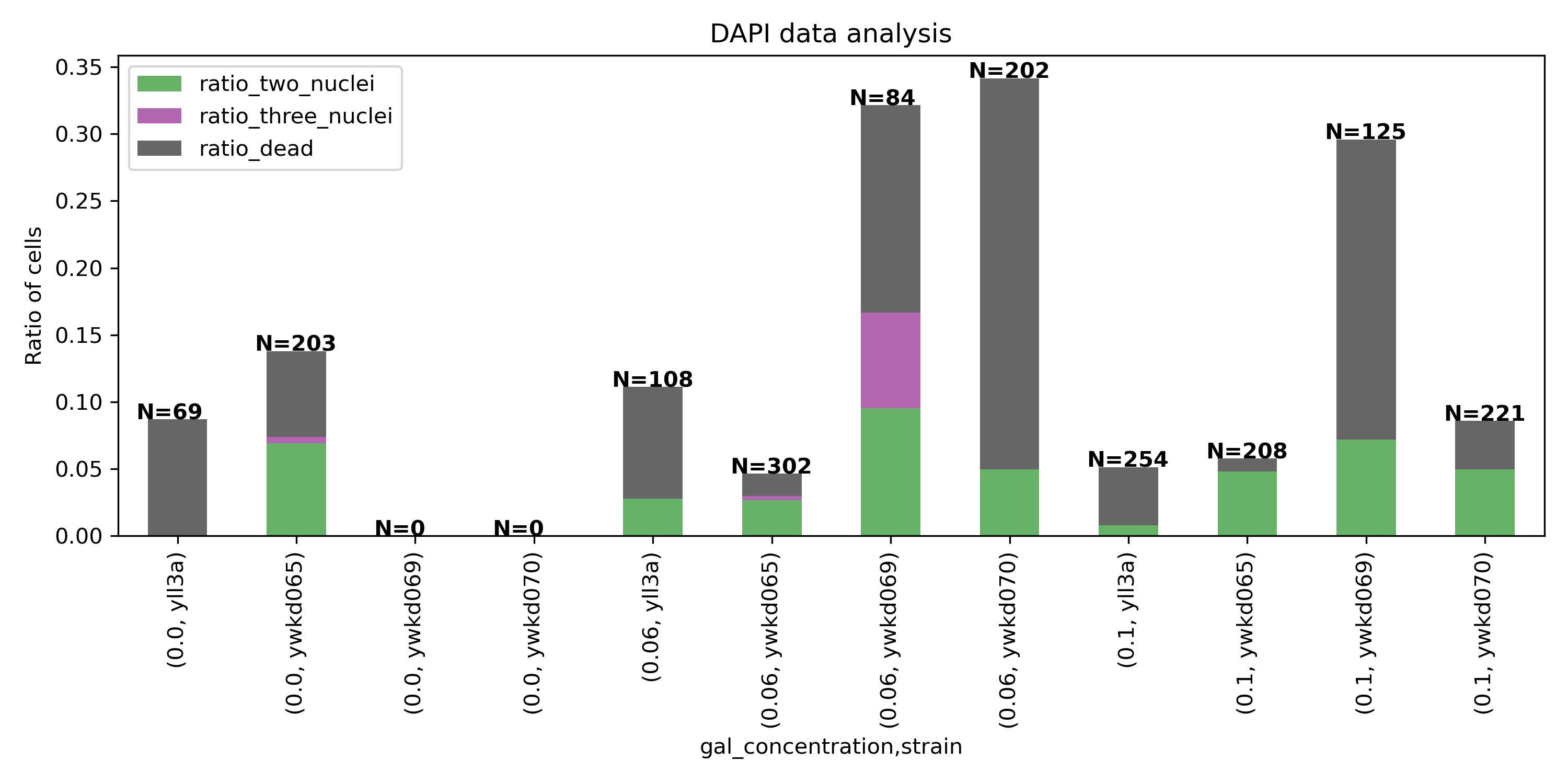
Check for multinuclei, and quantify the ratio of multiclei cells across the different conditions.
We expect an increase in multiclei cells in the dbem1 mutants compared to dbem1dbem3 mutants, and compared to WT.
7.4. Results#
11102022: Incubation in 30C at 10:30am
17102022: End of measuring in 36C + 48h incubating in the biotek
7.4.1. DAPI staining#
50ul in 100ul EtOH for 1h at room temperature
Spin dpwn at 2500 rpm for 1 min
Resuspend in 1ml 1xPBS
Spin down at 2500 rpm for 1 min
Remove the supernatant
Prepare a DAPI mixture: 0.5ul DAPI in 1ml 1xPBS (1:2000 dilution)
Bring all the eppies from all the cultures to the microscope and prepare the slides on demand for imaging due to the quick time of absortion and bleaching of the DAPI.
I had an issue in the microscope with the settings of the Z stack acquisition, since I using the “Multipoint acquisition tab” which was measuring in random locations that I did not select. I had to use the “ND acquisition tab” instead.
7.5. Data analysis#
The data analaysis was performed with the Image J plugin the Cell Counter1. Each cell was labelled to count the total number of cells in each frame and then each cell with two , three and dead cell was labelled as a different type.
The resulst were exported to a csv and analyzed in python.
7.5.1. Examples#
Two nuclei cells
-1.png) -BF.png)
Three nuclei cells


Dead cells