Title : 01102019- II pBK549 transformation on ylic133 for further sanity check in SATAY :pensive:
Contents
65. Title : 01102019- II pBK549 transformation on ylic133 for further sanity check in SATAY :pensive:#
65.1. Date#
01102019-03102019
65.2. Objective#
To ensure that the constructed strain is able to pass the Satay sanity check, and then I can continue with the further steps, like mating with yEK7a.
65.3. Method#
30092019 -Incubation of single colonies from the plates from 24092019 and Byk832 as positive control from glycerol stock at 10:30.
01102019 - OD measurements at 9:35
OD-10X dilution |
Titer |
Dilution factor to OD=0.5 |
Time |
|
|---|---|---|---|---|
ylic133_1 |
1.214 |
12.14 |
24 |
at 9:35 |
ylic133_4 |
1.216 |
12.16 |
24 |
at 9:35 |
ylic133_5 |
0.316 |
3.16 |
6 |
at 9:35 |
Byk832 |
0.308 |
3.08 |
6 |
at 9:35 |
at 9:55 Incubation at OD~= 0.5 to OD~=2 in YP+6x ADE
OD-10X dilution |
Titer |
Ready to transform |
Time |
|
|---|---|---|---|---|
ylic133_1 |
0.121 |
1.21 |
close to OD=2 |
at 13:15 |
ylic133_4 |
0.118 |
1.18 |
close to OD=2 |
at 13:15 |
ylic133_5 |
0.233 |
2.33 |
yes |
at 13:15 |
Byk832 |
0.290 |
2.9 |
yes |
at 13:15 |
at 14:30 30C incubation
100ng of pBk549
Notes on the procedure :
I did not vortex the cells with the transformation mix, instead I pippeted in and out after each ingredients, by this way I avoided clumps formation that hinder transformation efficiency.
I did not use the maximum speed (13300) of the centrifuge to get rid of the LiAc, instead I used 8000rpm.
I heated the ssDNA to 95C and then transfered to ice before I added the 0.1M LiAc , so I could do all steps of LiAc washing and samples separation consecutively.
I plated all (200ul) in the selection plates (-URA+6x ADE), both, negative control and transformed cells.
65.4. Results#
031022019 No clonies in the selection plates :pensive: I just see 3 colonies in ylic133_1 plate.
Possible causes:
The plasmid is degraded
check: I loaded 10ul of plasmid into a gel to see If I have low bands, that could indicate degradation of the plasmid.
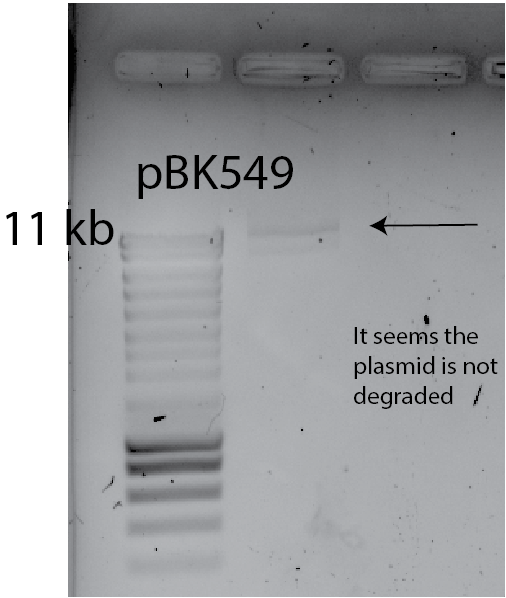
This result implies that plasmid degradation IS NOT THE CAUSE.
The plasmid was lost
check: I restreak one colony I had on the selection plate into a new -URA+6xade plate to see if they actually acquired the plasmid:
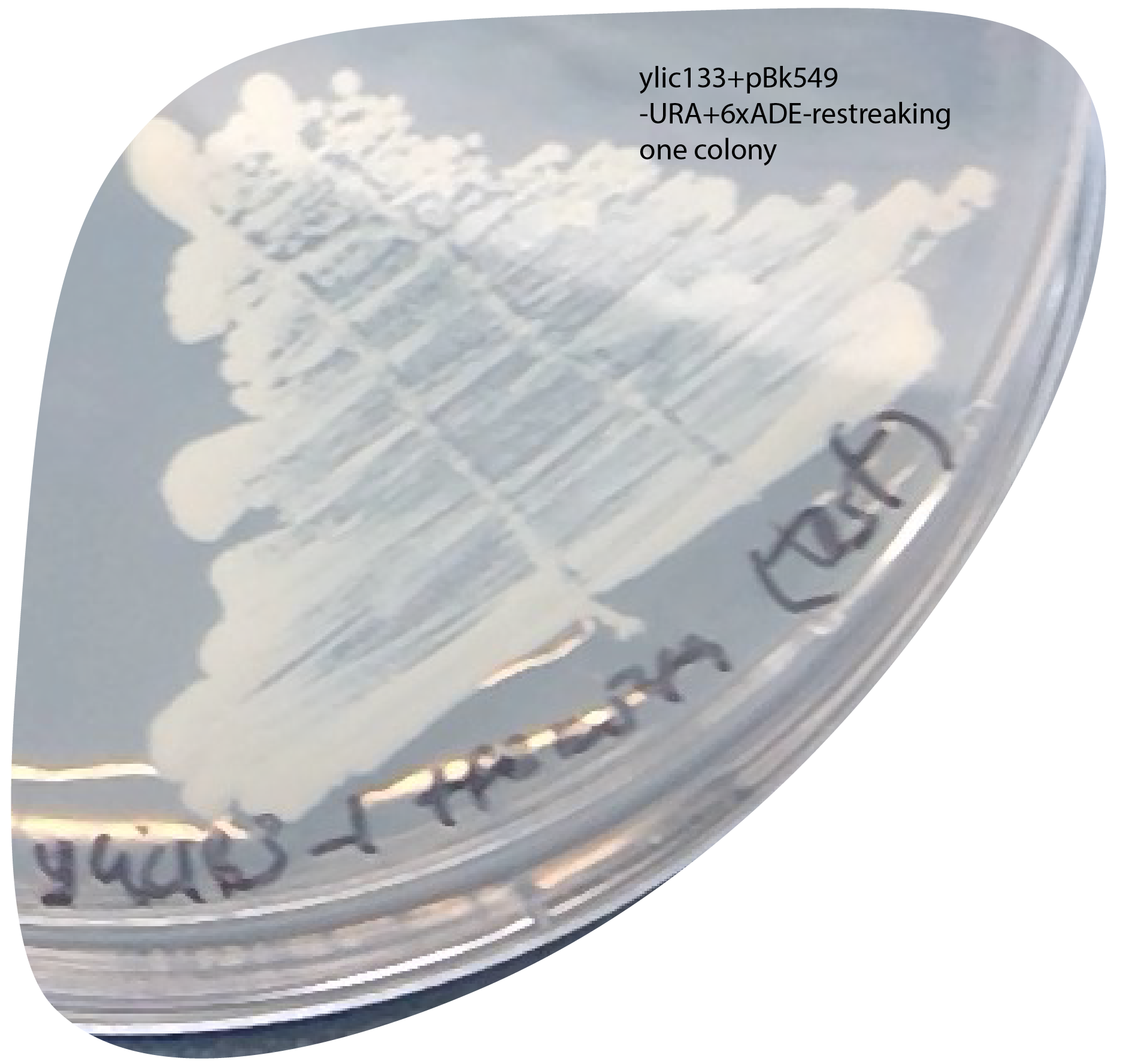
This implies that is neither the cause.
I used an extremely low amount of plasmid
This could be due that I did not vortex the plasmid before using it, and maybe some of it sediment to the bottom, and effectively I took less plasmid than what I thought. Next time I should vortex it anyways.
The selection plates are not good i.e. nothing can grow there.
check: I plated a positive control (yWKD017 and ylic132) on the -ura plate+6x Ade I used for the selection plate.
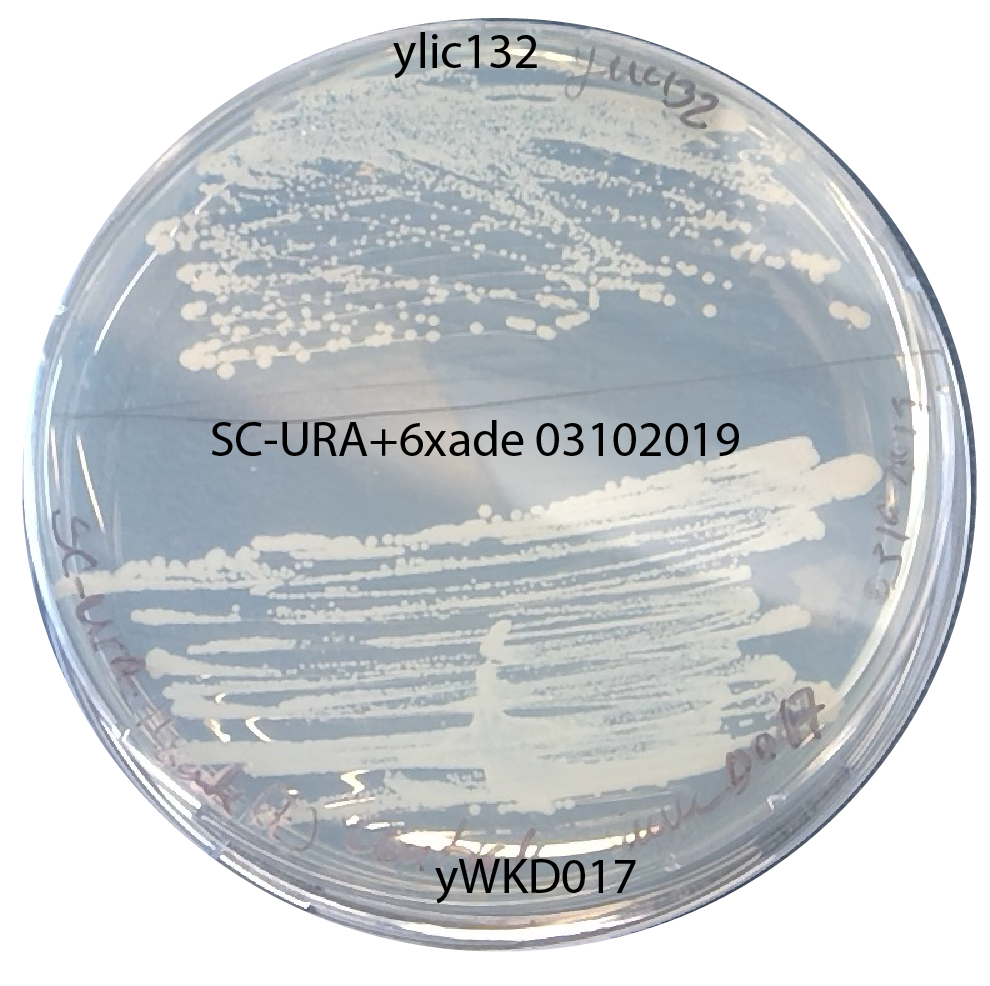
This growth of positive control on the plate indicates that the plates ARE NOT THE CAUSE
My reagents of the transformation protocol are not good.
Maybe my PEG50% is not well… If the next transformation does not work , I will replace all my reagents for new fresh ones.
I actually replace the LiAc , I did a new 1M stock of 50mL.
65.5. Conclusion#
Still I dont know the cause of why my transformation arent working, though I discarded multiple causes.
I will try again with new plasmid from another bacteria incubation, and after checking that my plates are indeed correct, to discard those factors.
