Title: Transformation of ylic133_5 with the PCR products from yll137 and yll140
Contents
54. Title: Transformation of ylic133_5 with the PCR products from yll137 and yll140#
54.1. Date#
23022020-
54.2. Objective#
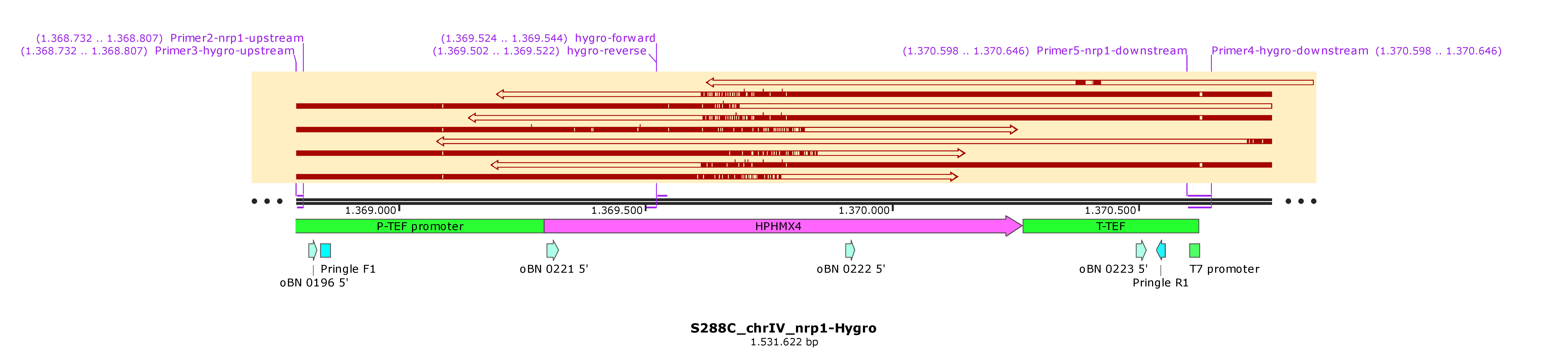
Transformation-1: To replace the nrp1 locus with the HYGRO cassette using the PCR product of yll137.
Transformation-2: To replace bem2 locus with the leu2 marker using the PCR product of yll140.
54.3. Method#
Following this Yeast Transformation Protocol
Day0: 16:00 , Incubation of ylic133_5A from glycerol stock from 01202019 in YPD+6xADE. 10mL per transformation with its negative control. 6xADE=44mg/400mL =110mg/L , Benoit uses 40mg/L
Day1: Measure OD of overnight cultures
Sample
10X dilution OD
Real OD
yll133_5 (1)
0.1
1
yll133_5 (2)
0.07
0.7
At 10:00 am 2nd incubation with a 2x dilution down to OD~0.5
Check at 14:00 the OD
Sample |
10X dilution OD |
Real OD |
|---|---|---|
yll133_5 (1) |
0.28 |
2.8 |
yll133_5 (2) |
0.23 |
2.3 |
DNA amount for transformations
nrp1::HYGRO - 806 ng (73.3ng/ul*9ul)
bem2::leu2 - 1.296 ug(144.4 ng/ul*9ul)
The plates to check the success of the transformations are:
YPD+HYGRO+6xADE
-leu2+6xADE
The only autoclave component was the agar (4g) with 100ml MiliQ, the rest was added by filtering (in 80ml MiliQ). To avoid degradation of aminoacids in the autoclave machine.
Plating:
ylic133_5 + bem2::leu2 , low and high concentrated , 90x and 10x dilution, for a final volume of 200ul.
Recovery step for ylic133_5+nrp1::HyGRO
After removing the transformation mix , I added 1ml of YPD+6xADE to incubate in 30C overnight, and then plate that next morning on the selection plates.
This choice for the recovery way is based on the experience when selecting using 5FOA plates for not having URA, here.
54.4. Results#
25022020- Samples were spilled out from the eppi tube after the overnight incubation (I put too much YPD that cause the burst due to CO2 production)
Next time put parafilm, around it , to prevent spilling.
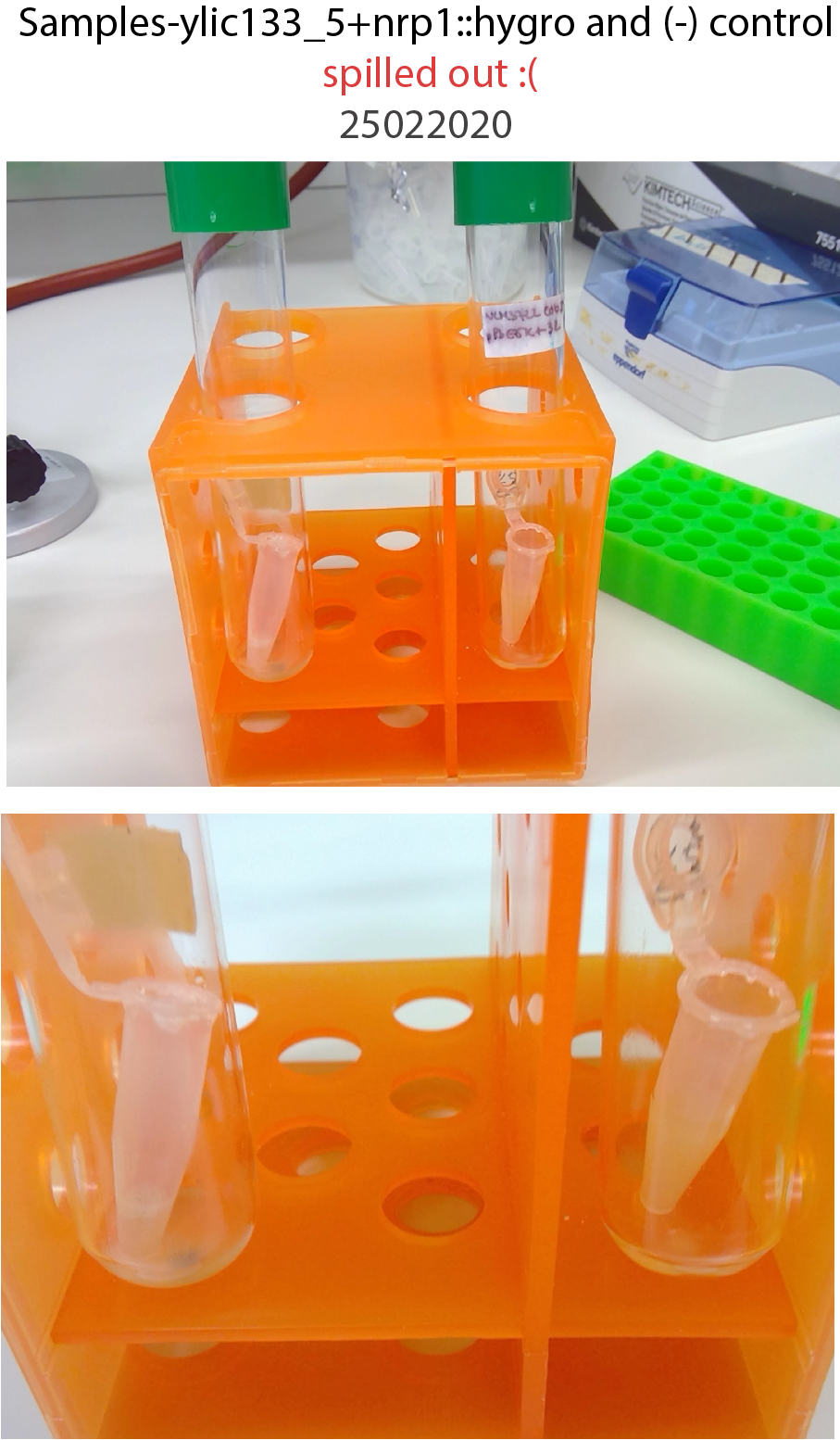 {width=50%}
{width=50%}
Note:
In general it is not advisable to wait more a whole overnight for recovery, because then we decrease the changes to have different biological replicates, since maybe one population will take over during the overnight incubation.
Esentially the stadistics will go down , and if we have suppresor they will be “hidden” when plating on the selection plates.
I got more than 50 transformants in the -leu2 plates and a big splash of cells combiend with colonies in one of the hygro plate.
I inoculated 200ul from the eppi tube that spilled out, onto the two plates, without any dilution.
In one of the plates there was a big loan because they were many colonies on it.
I did not see any bacterial contamination on the plates. :)
Colony PCR to check the transformation
I check 8 colonies from the hygro plate and -leu2 plate and retreak them onto a new plate, to save the colony.
First colony PCR I did using SPY1/2 Els primers and 1ul of template.
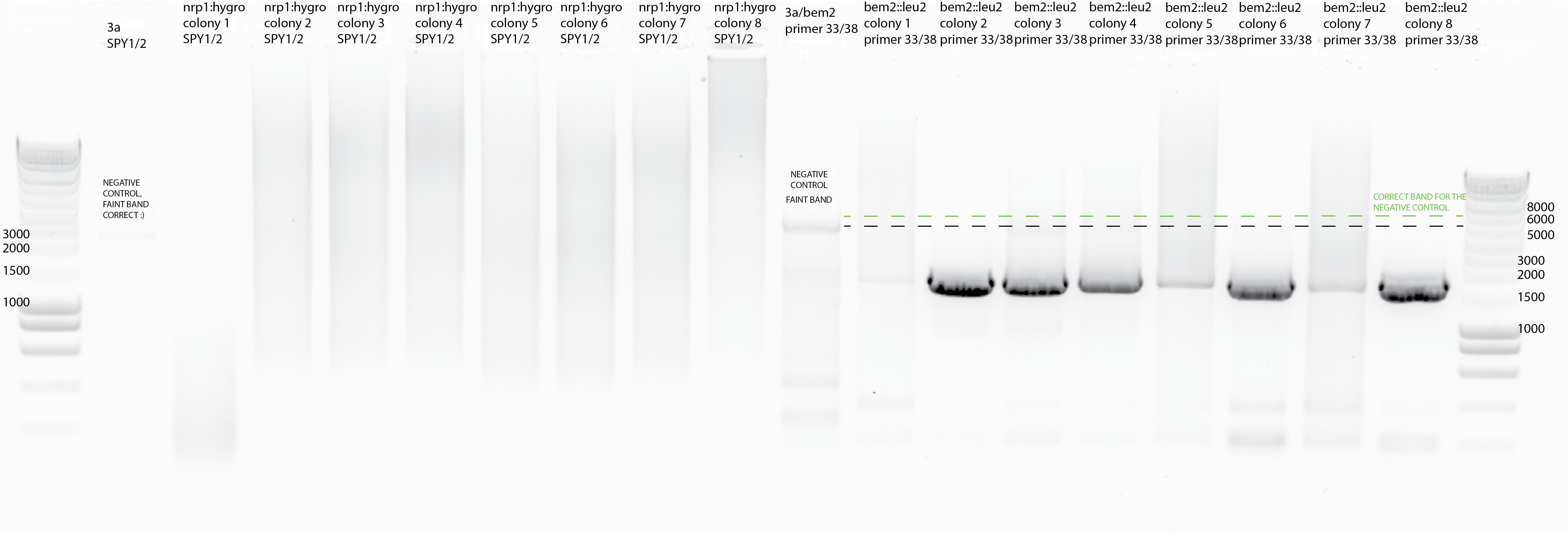 {#fig:colony_pcr_1}
{#fig:colony_pcr_1}
 {#fig:colony_pcr_2}
{#fig:colony_pcr_2}
For the nrp1::hygro clones it has not worked with SPY1/2 primers.
Colony PCR for nrp1::hygro clones with 27/32 primers from my stock and 83/84 from Els.
I take single colonies from the restreak onto Hygro.
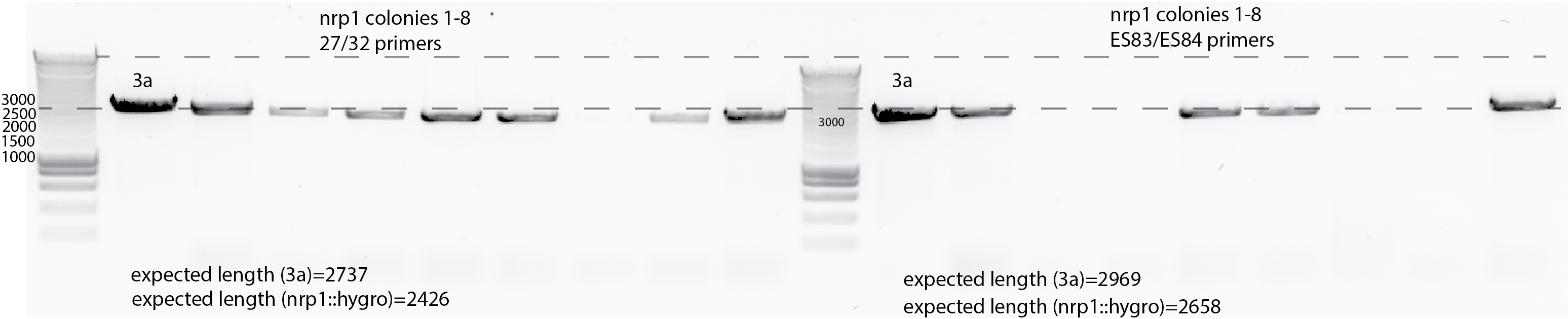
It is not clear whether the nrp1::hygro have the right length. Neither the negative control with primers 83/84.
Do another PCR with sequencing primers to check again. 03032020
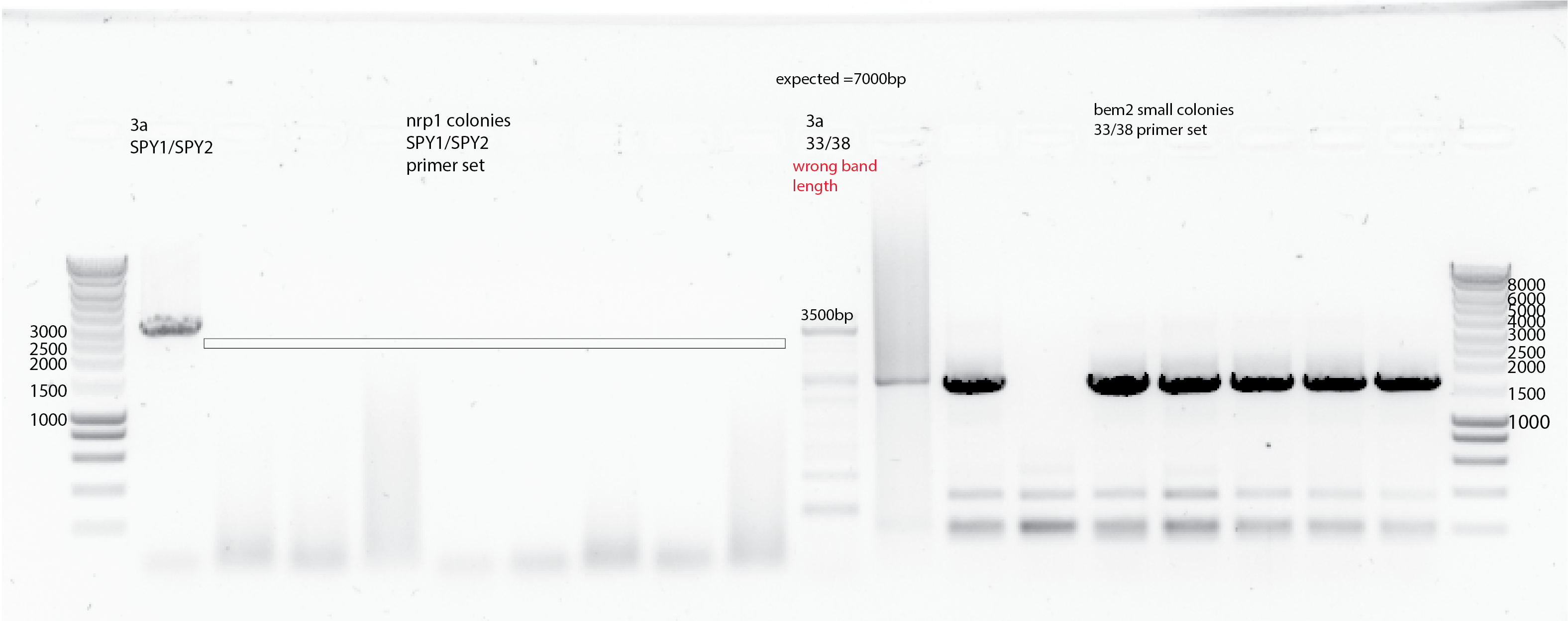
I did the PCR with the sequencing primers and redo the gel for the previous PCR with primers 27/32 and 83/84 but running the gel 40 mins at 110V. (Following Els and Liedewij recommendations from lab meeting). Below are the results:
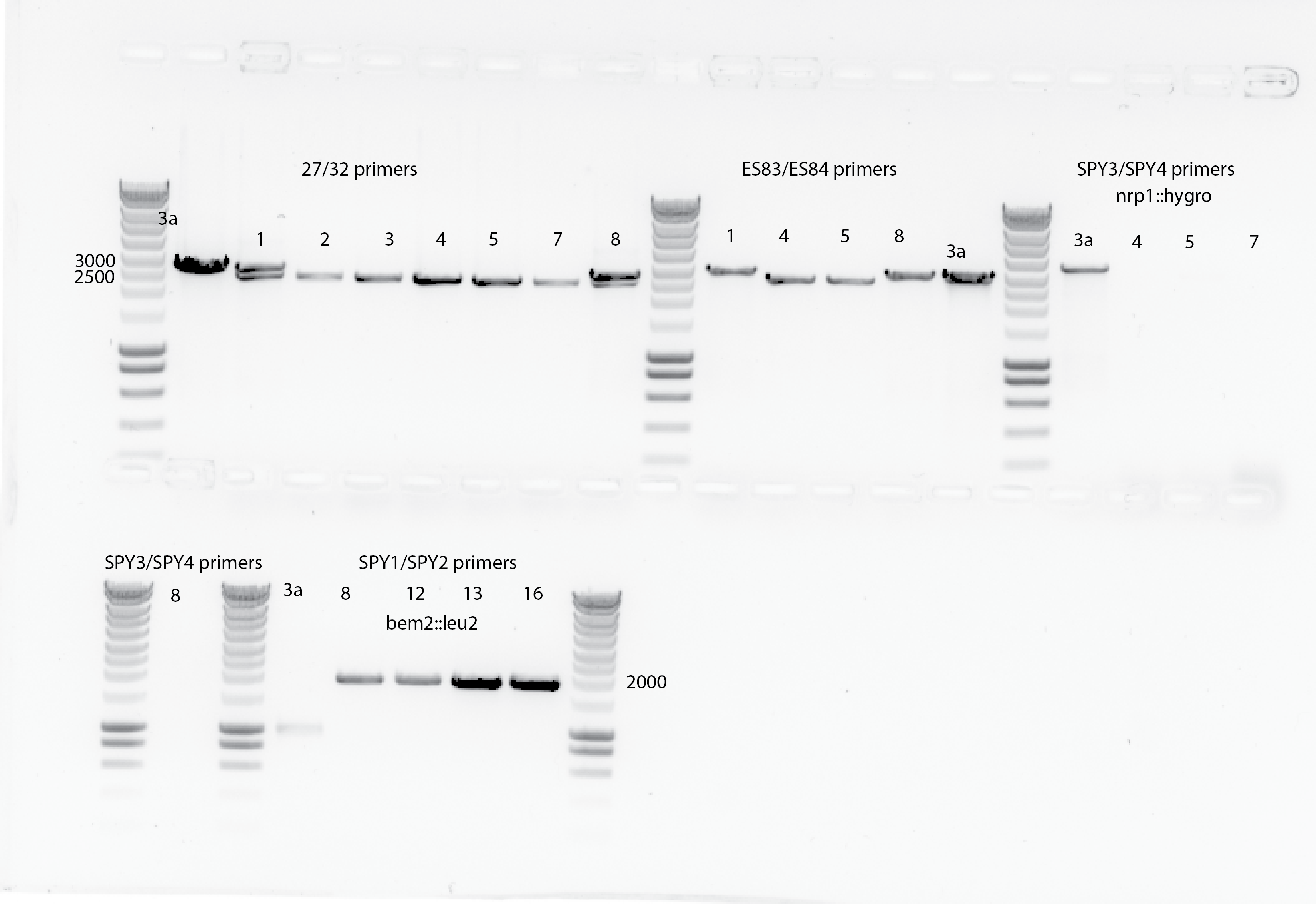 {#fig:seq-PCR}
{#fig:seq-PCR}Results:
Colony 1 and 8 from nrp1::hygro clones seems have both(?), and the rest seems OK according the band lenght. This observation is also consistent with primers 83 and 84. No bands are shown for the sequencing primers for the nrp1::hygro clones.
For bem2 it seems also OK, all the bands. Still yll3a does not show the right band in 7000bp which could be due to the PCR cycle.
I took single colonies (8,12,13,16) from the -leu2 plate restreak and grow them in YPD+6xADE for glycerol stock and restreak them in normal YPD plate to check for the red phenotype.
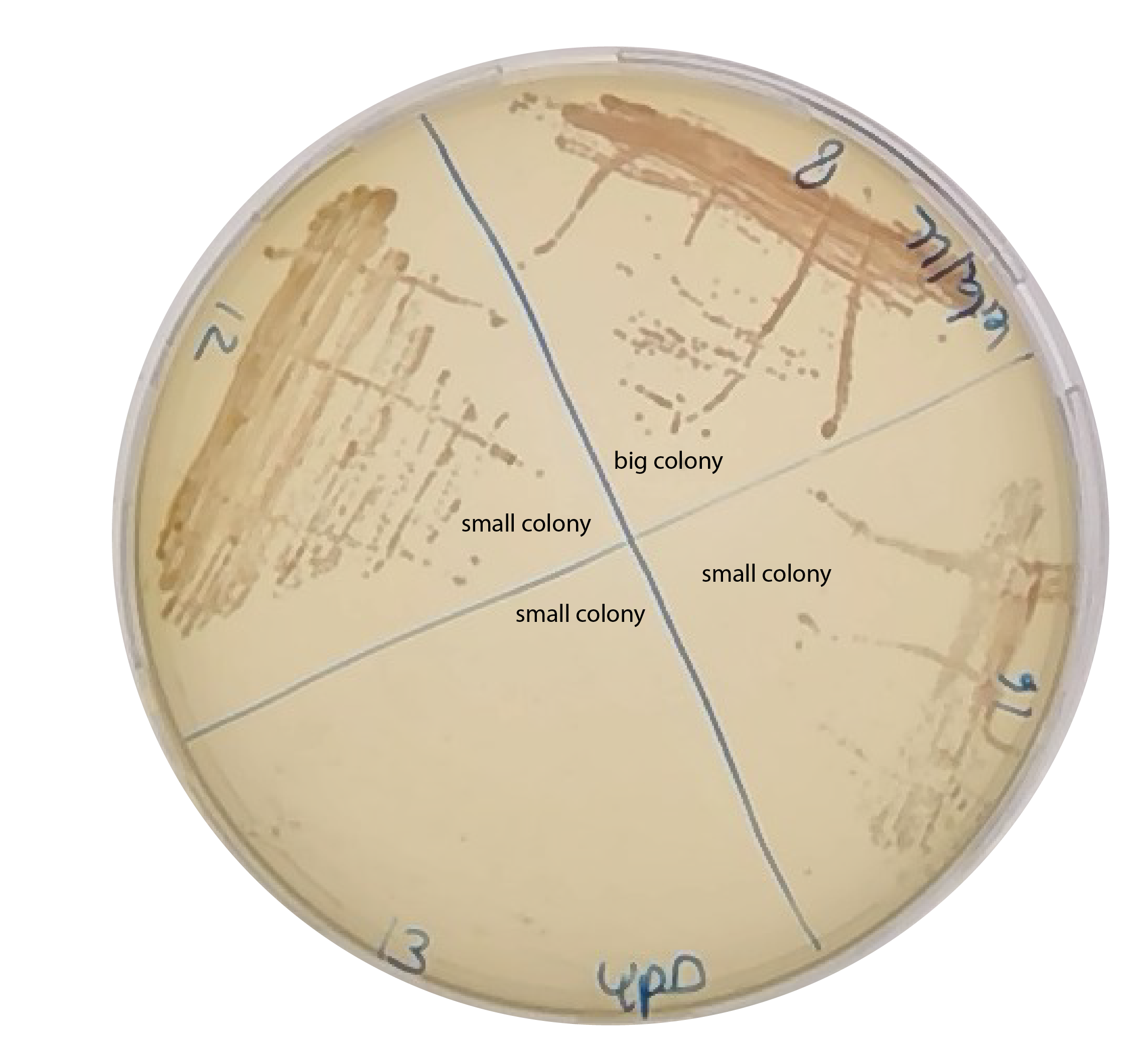
Do colony PCR with SPY1-bem2 and SPY2-bem2 with all small and big colonies that have shown a band with 33/38 primers.
Store also 5 small and 5 big colonies to later test the population phenotype, and decided which is bem2d. Because it seems I have two populations, that show the right band on gel.
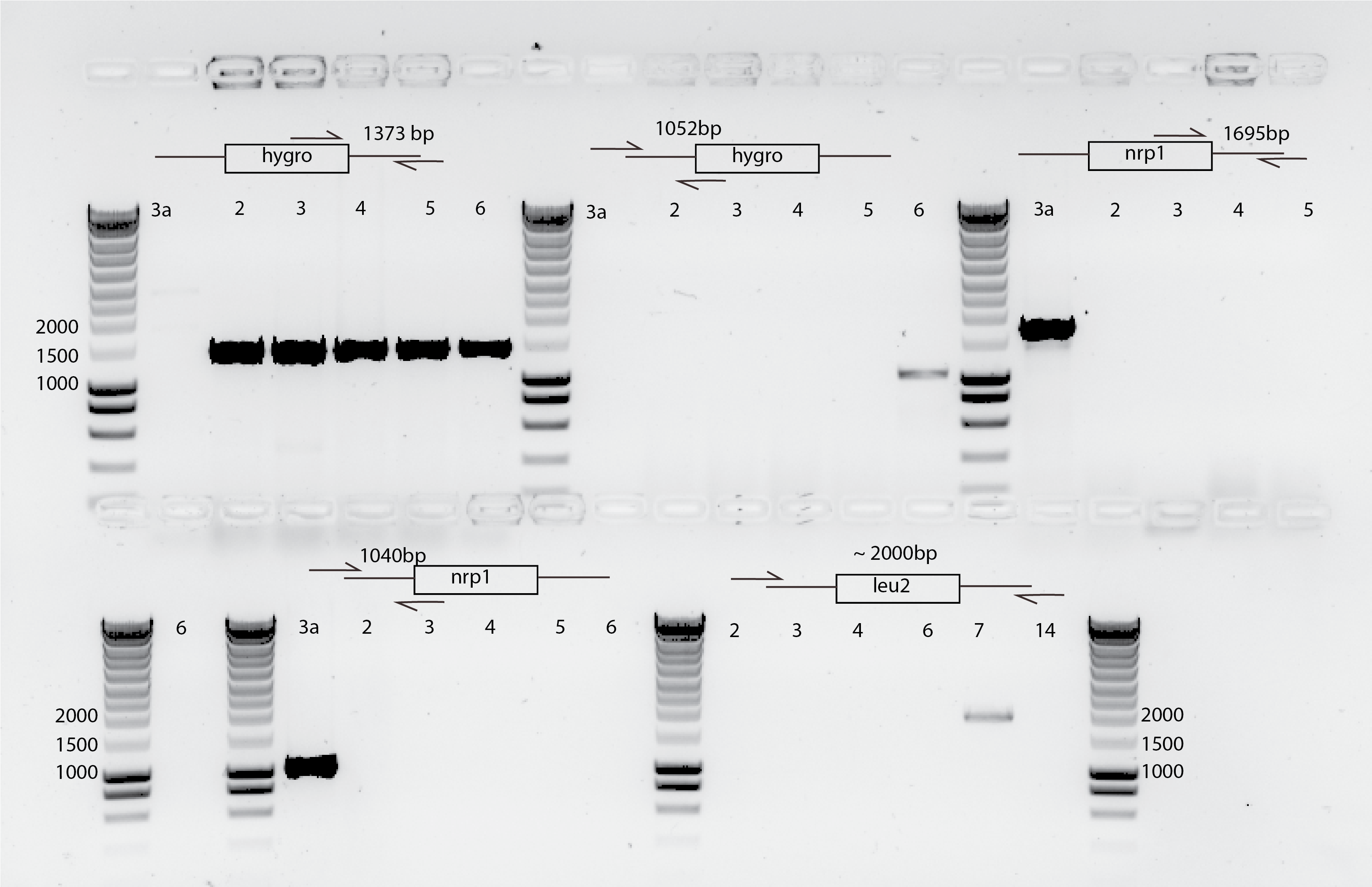
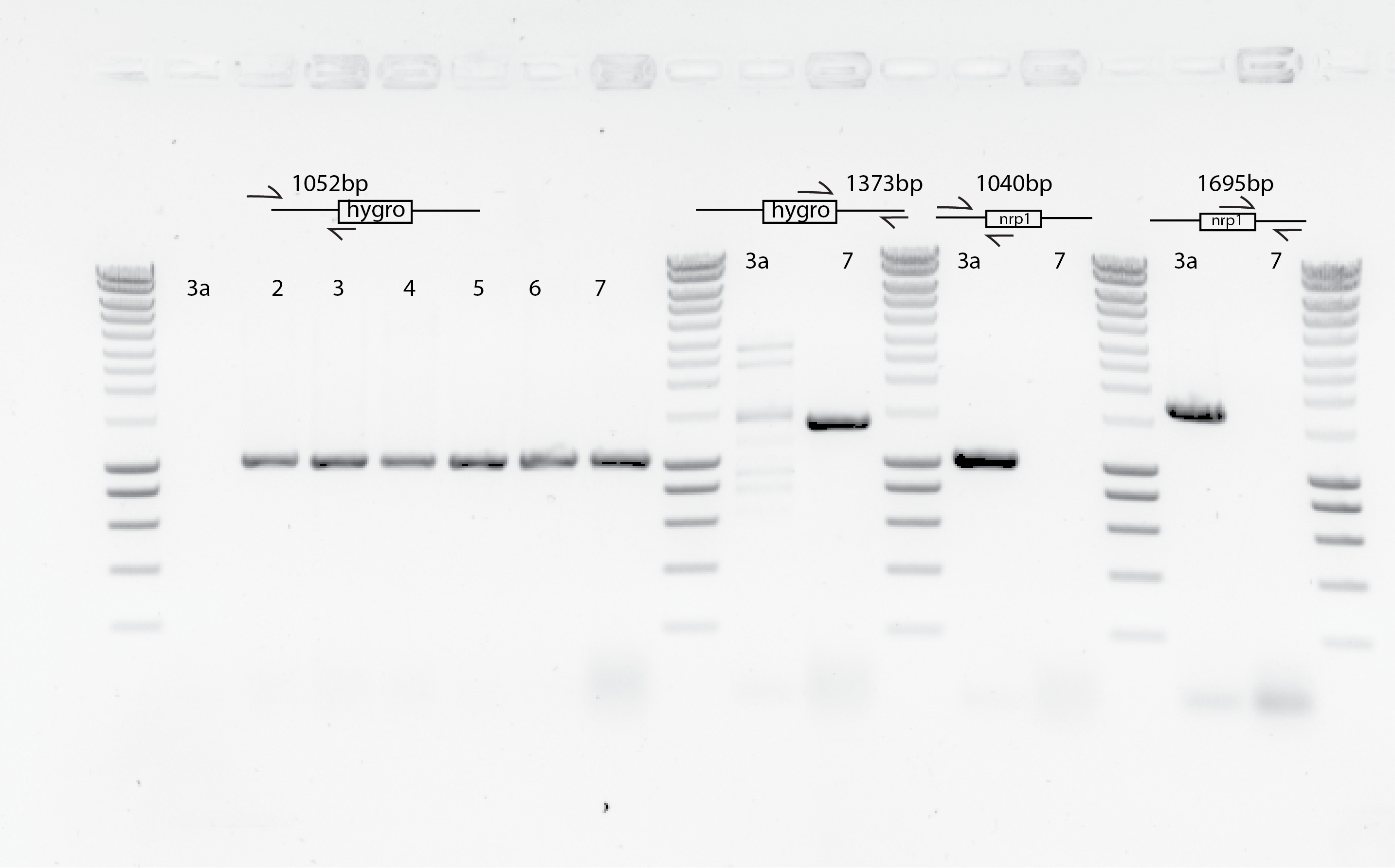
54.5. Conclusion#
It seems I could take 5 clones per transformation that seems they are correct :)
I sent already (05032020) bem2::leu2 clones to sequence with primers SPY1 and SPY2 (good results)
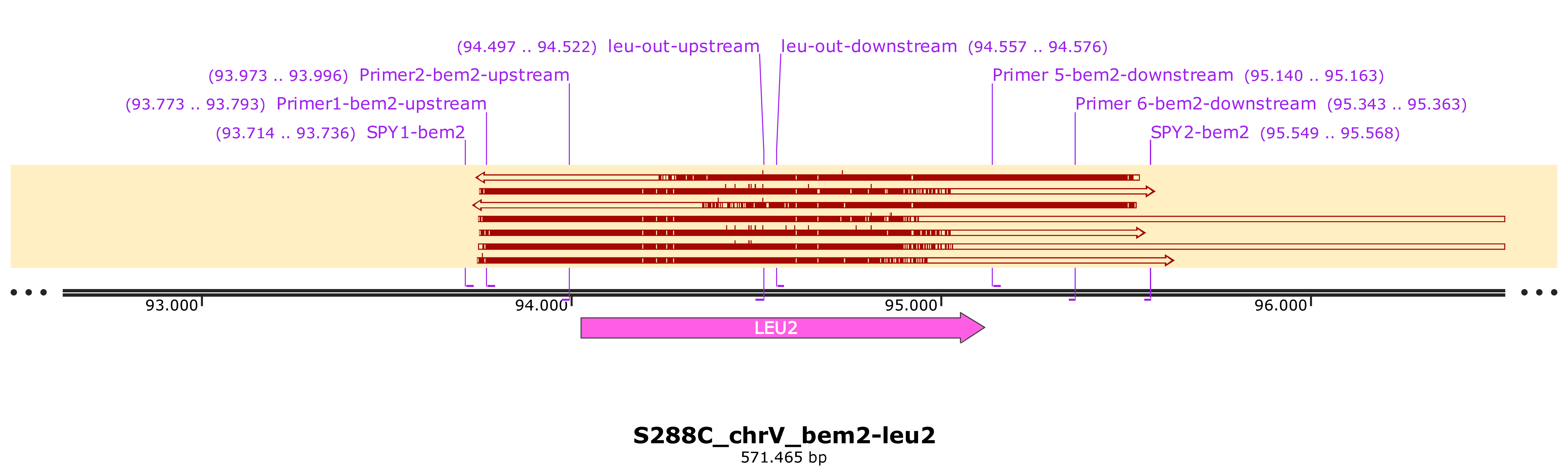
I will send today (06033030) nrp1::hygro clones to sequencing.
I am also incubating those clones in liquid selection media to do a population phenotype test in the Biotek. Results :)
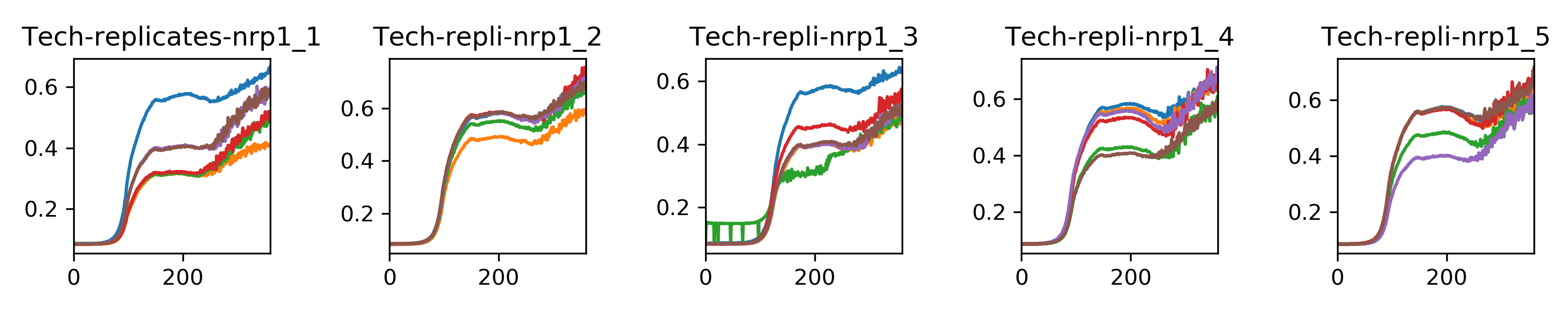
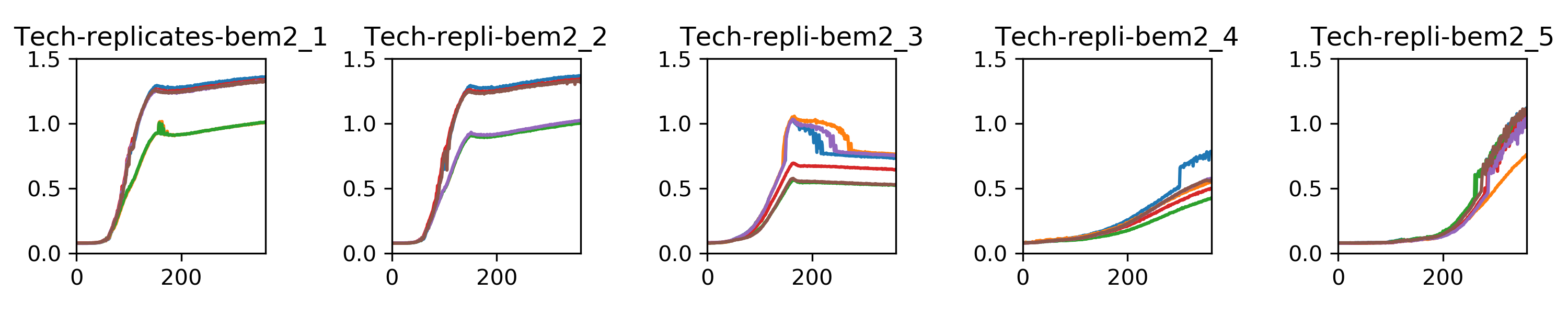
The code I wrote to compute the doubling times is in this jupyter notebook
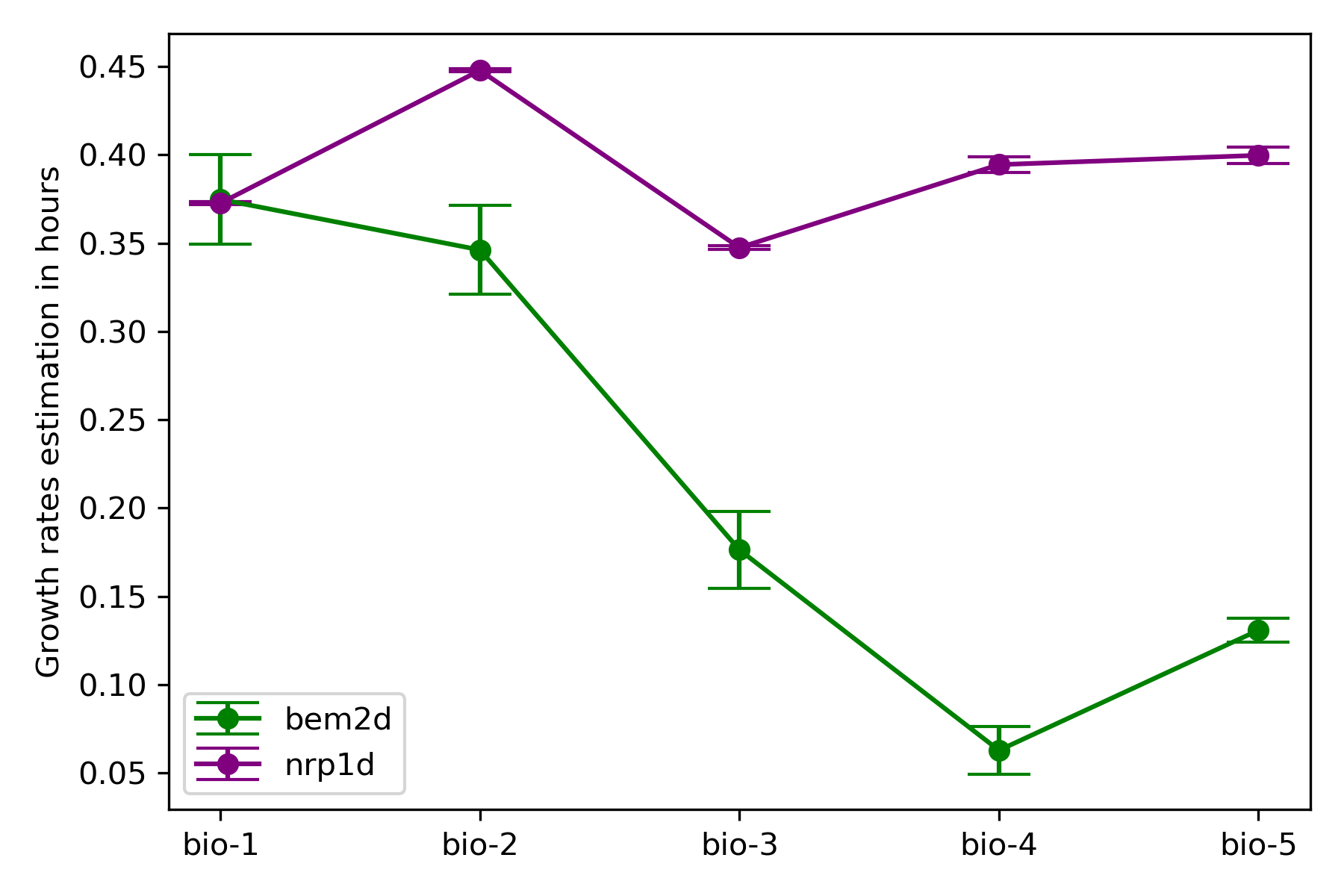 {width=60%}
{width=60%}Indeed there is less variation among nrp1d clones than bem2d clones, which could be due to the overnight recovery.
I will also stored them today as glycerol stocks in the selection media.
I stored only one tube per biolog replicate.
54.6. Next Steps#
Repeat the biotek measurements using the wt strain in order to compare these mutants relative to the WT.
Make a PCR from inside the leu2 marker and bem2 to check the correct integration of bem2::leu2 transformants. (colony PCR) still is not clear the differences I see in the selected biological replicates for bem2::leu2 mutants
Primers ordered and already arrived :)
