Title : bem1::KanMX transformation in WT+pGal-CDC42
Contents
22. Title : bem1::KanMX transformation in WT+pGal-CDC42#
22.1. Date#
06042021-23042021
22.2. Objective#
To have this strain for the growth controls measurements needed for the paper with Fridtjof.
The strain we had yIdb005a,b are completely contaminated with WT. See the last results from 2021-01-25-Supp-controls-sfgFP-influence-to-paper
22.3. Method#
Yeast Transformation Protocol
06042021: Incubation of ywkd071a in SC-URA+2%Gal+2% Raff
07042021: Culture not in log phase, only at late afternoon , around 15:00. I transfered the culture to room temperature to do the transformation on the next day.
08042021: OD measurement
strain |
OD 10x dilution |
Real OD |
Dilution to OD=0.5 |
Real OD after dilution |
|---|---|---|---|---|
ywkd071a |
0.7 |
~7 |
10x |
0.4 |
Incubation started at 9:00
Start of transformation at 13:00 , with OD=1.3
10ul of DNA from PCR bem1:KANmx (~200ng/uL) = 2ug DNA
2h recovery step in SC-URA+2% Gal+2% Raff
Plate in selection plates (Sc-URA+2%gal+2%raff +G418)
100ul 0x dilution (positive control)
100ul 50x dilution (positive control)
100ul 100X dilution (positive control)
100uL 0X dilution (negative control)
Plate in normal plates (SC-URA+2%gal+2%raff )
100uL 100X dilution (positive control)
100uL 100X dilution (negative control)
22.4. Results#
Selection plates
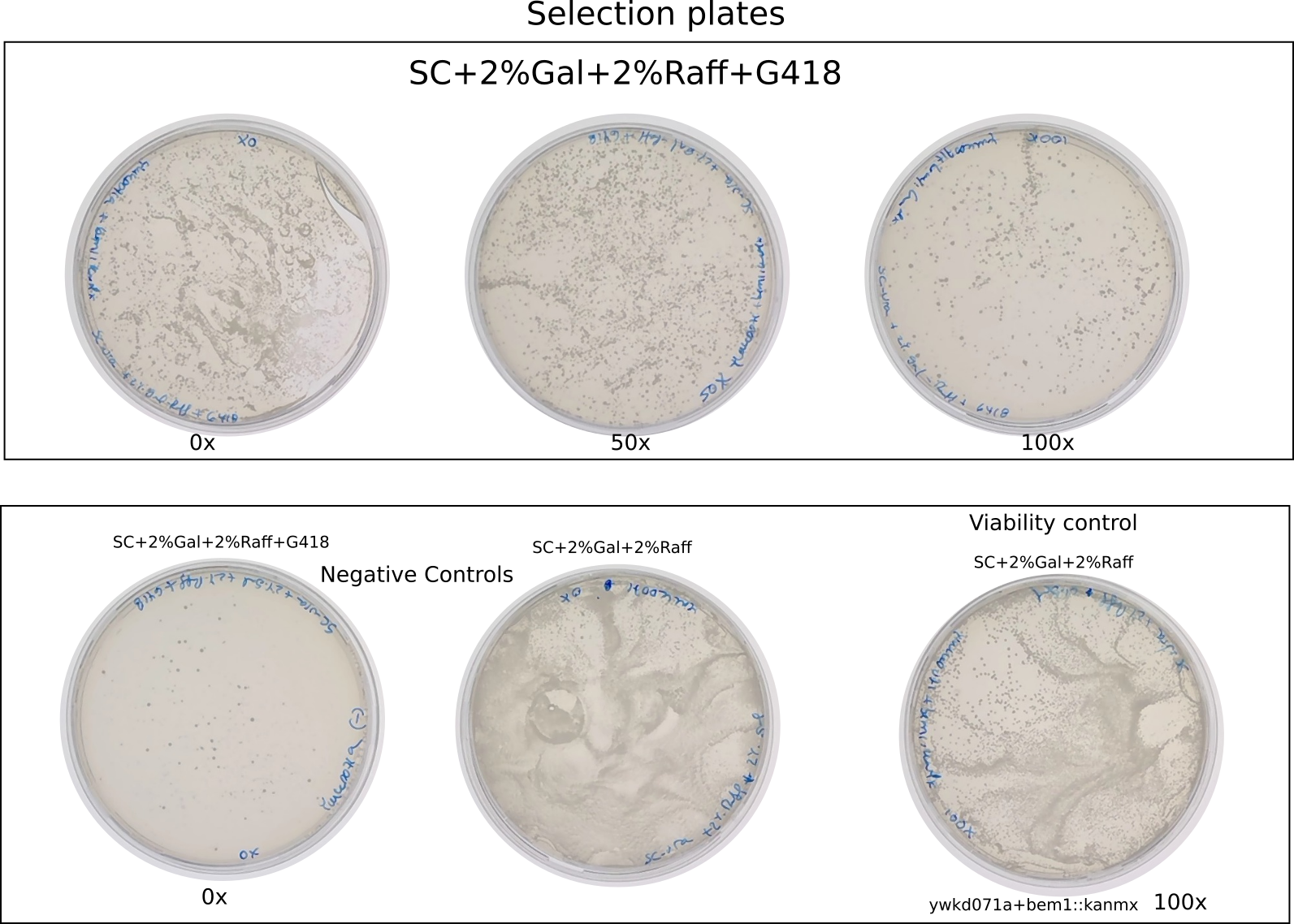 {#fig:selection-plates}
{#fig:selection-plates}
Colony PCR with 4 from small to medium size colonies
Primer sets: olic56/oll60, olic57/oll61, olic56/olic57
Colonies dissolved in 25ul MiliQ
1uL for PCR
PCR with Leila_LL_60 protocol : 60C annealing temperature, 2min in 72C and 30secs in 98C
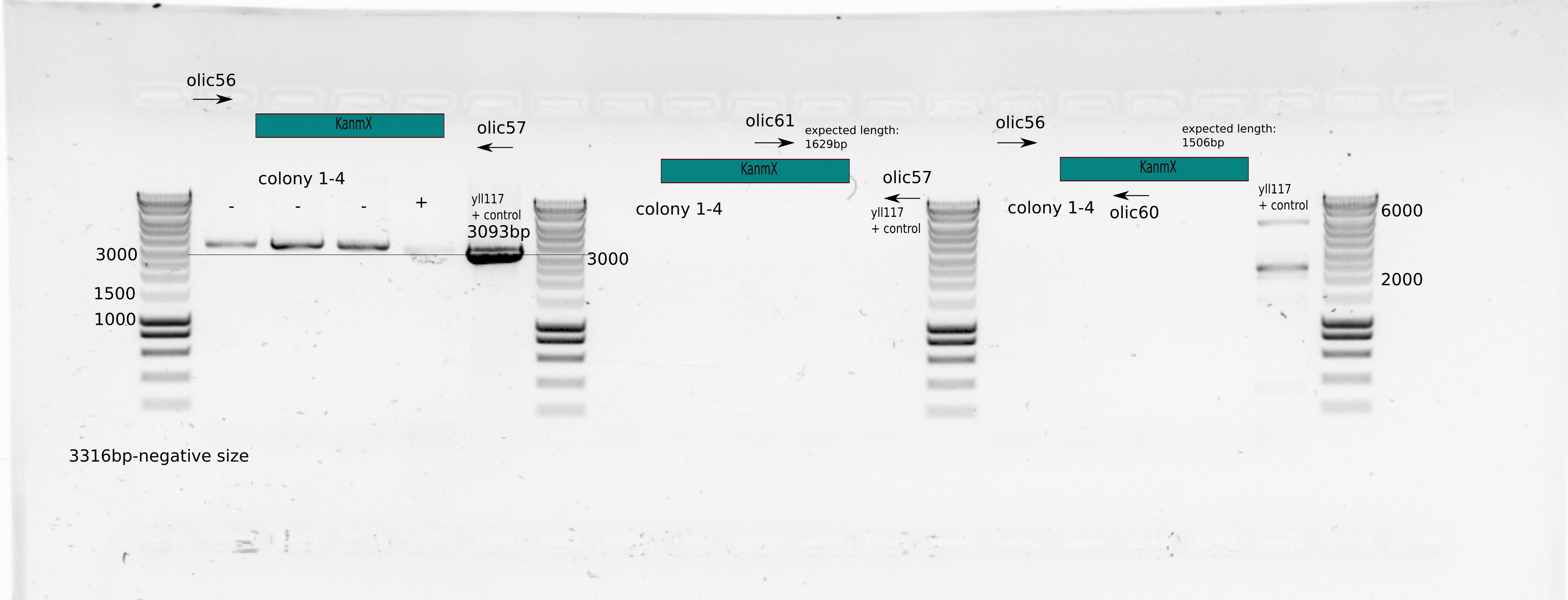 {#fig:colony-pcr}
{#fig:colony-pcr}
We can not say anything from Figure [@fig:colony-pcr] because it seems the primers oll60/61 dont work. It seems that colony 4 could had a band size for the external primers similar to the positive control, but it did not grow in plate.
Colony PCR of more colonies , one of them a big colony from the 100X selection plate.
Primer sets: olic56/oll30, olic57/oll29, olic56/olic57
Colonies dissolved in 10ul MiliQ
Take 1ul for PCR
PCR with Leila_LL_60 protocol : 60C annealing temperature, 2min in 72C and 30secs in 98C
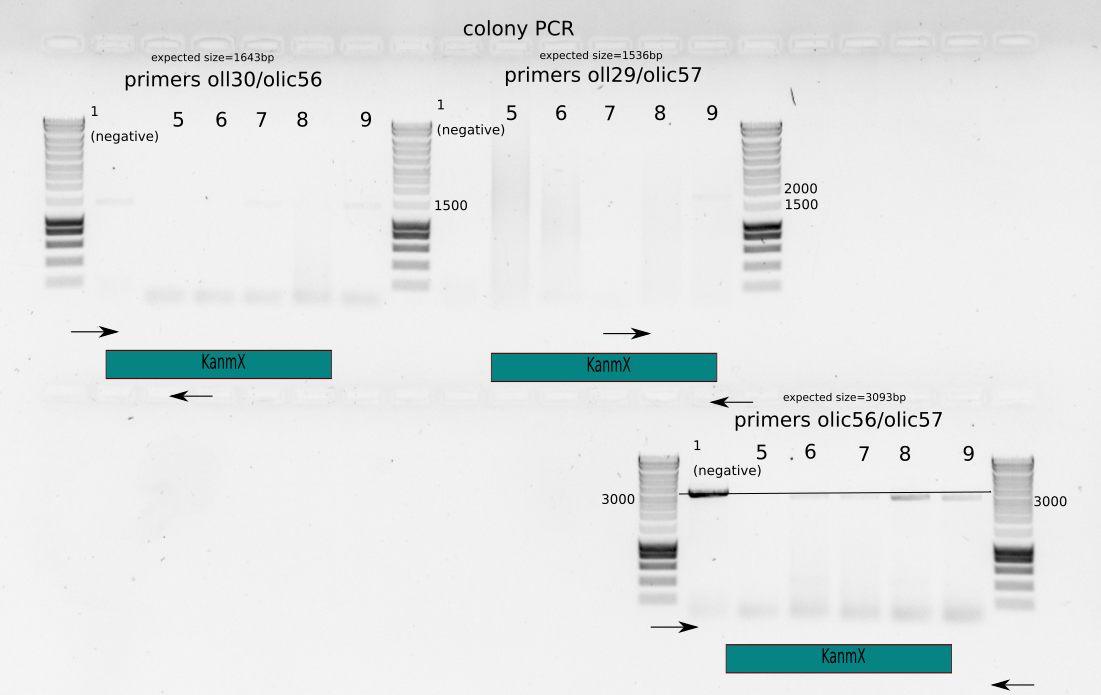 {#fig:colony-pcr-colony-9}
{#fig:colony-pcr-colony-9}
From Figure [@fig:colony-pcr-colony-9] we can see that colony 9 shows all the exected band sizes. Though it is very vague for the edge selection.
Put all the colonies to grow in SC-URA+2%GAL+2%Raff+G418.
Colony PCR with two more big colonies from the 100x selection plate and two more from the a re-streak plate.
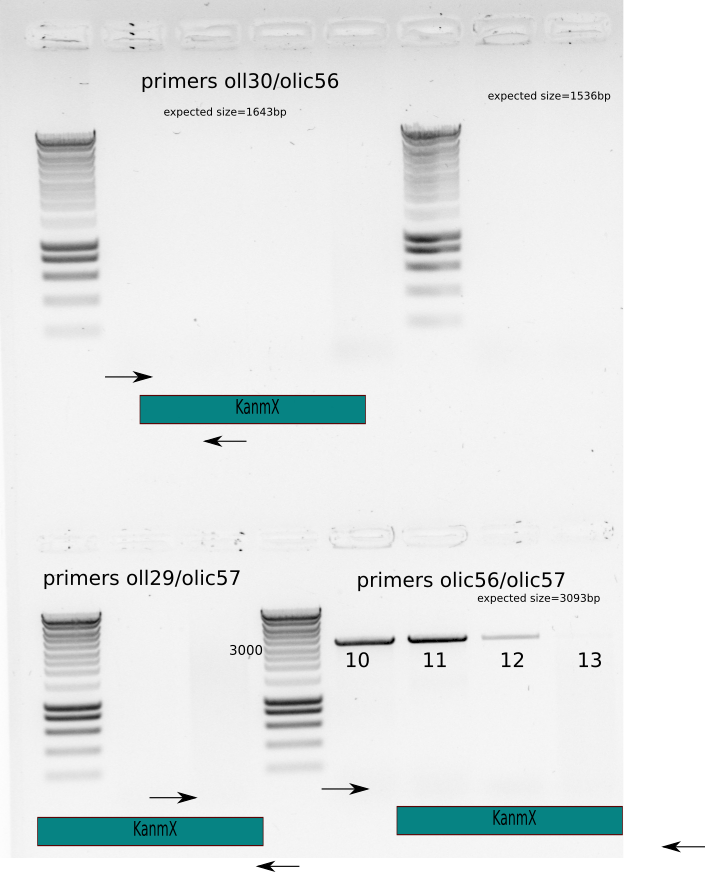 {width=50X}
{width=50X}
could be because of the colony PCR
19042021-Liquid culture of colonies 10,12,13(they were like dehydrated, not wet colonies in the plate),and 9 , they were the ones that shows growth over-weekend in plates.
Media: SC-URA+2%Gal+2% Raff+G418
gDNA extraction from these colonies and glycerol stocks
PCR with primers oll29/57 , oll30/56 and olic56/57
Storing three biological replicates for glycerol stocks.Colony 9 did not grow . Strain name : ylic139
colony 10: a
colony 12: b
colony 13: c
 {#fig:pcr-with-gdna}
{#fig:pcr-with-gdna}
Results from the Figure [@fig:pcr-with-gdna]
replicate ylic139 c shows a weird band size for the upstream and correct integration. So it seems it does not have a correct integration.
yll3a shows a band for the ptEF terminator , which should not be possible.
The rest of the replicates seem fine.
22.4.1. Positive and negative checks for the glycerol stocks#
Negative controls
Plate the replicates in SC-URA+2% RAff+0% Gal, including yIdb005a (the contaminated stock)
Inoculate them in liquid culture in SC-URA+2% Raff+0% Gal, including yIdb005a (the contaminated stock)
Positive Controls
plate them in SC-URA+2% RAff+2% Gal+G418 including yIdb005a,(the contaminated stock)
22.4.2. Results#
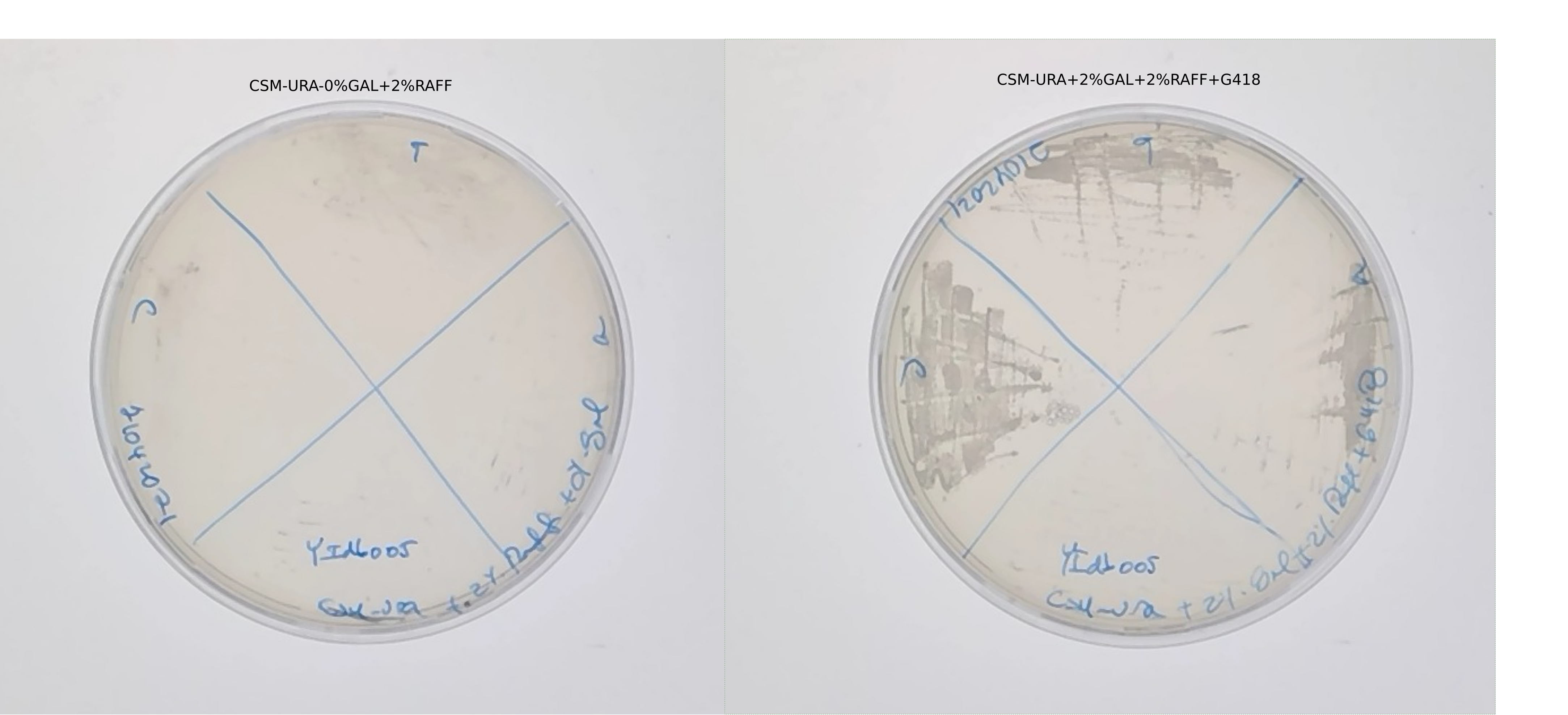
Growth of replicates from glycerol stocks in SC-URA+2%Gal+2%Raf+G418.
No growth of yIdb005 in selection media (confirmation)
Miserable growth after 5 days in 0% Gal of the replicates in plates.
Since the glycerols stocks are in 2% Gal they need to be dissolved in 0% Gal to dissolve the Cdc42.
22.5. Conclusion#
We have two biological replicates of new dbem1:KanMx pGal-cdc42::URA : ylic139 a, b .
