Title: FACs experiment with the same conditions as the microscopy done to quantify the cell sizes for the pAl1-Cdc42-sfGFP strains. (I)
Contents
60. Title: FACs experiment with the same conditions as the microscopy done to quantify the cell sizes for the pAl1-Cdc42-sfGFP strains. (I)#
60.1. Date#
20012020-23012020
60.2. Objective#
To be able to compare the results of the FACs with the microscopy conditions. And also to see if we get a difference in expression when increasing the incubation temperature to 36C.
60.3. Method#
Insights from the Gal1 promoter
The location in the genome at which the gal promoter was inserted can have a strong effect on the expression pattern of the gal promoter (Ramon feedback). Hence we should not compare different studies of the Gal1 promoter with ours if the integration in the genome is in a different location and also if it is a plasmid or not.
We should compare systematically the Gal1p expression pattern of the strains that has the sfGFP (Werner strains) and the mneonGreen ones (Ramon/Miranda strains), because they have the same type of genomic integration of the Gal1 promoter.
Ask Reza for his data with WT+mneongreen to compare with mine
Look for the postprocessing results, in this folder
Follow the same protocol as I followed for the microscopy measurements
 {#fig:experimental-design}
{#fig:experimental-design}
Planned procedure
15 hours of incubation in 2% Gal +2% Raff (i.e. overnight incubation from 17:00 to 08:00 )
Washing step with CSM+2% Raff+0%Gal to the respective Galactose (at 08:00-09:00) concentrations. Incubate
Measure FACs after 24 hours of incubation . (next day of the washing step)
Use the references cdc42-GFP ywkd038 and ywkd001 in 2% dextrose +2% Raff, in CSM-met and CSM respectively.
In order to have the same conditions as the microscopy done in December 2018 , where we quantify the cell sizes after 24 hours of incubation in X% galactose (after a washing step from a firsto overnight incubation in 2% galactose), the media has to have 4x the normal amount of aminoacids. This maybe has an impact on the regulation of the promoter.
The new plate design for this is shown in @fig:plate-design
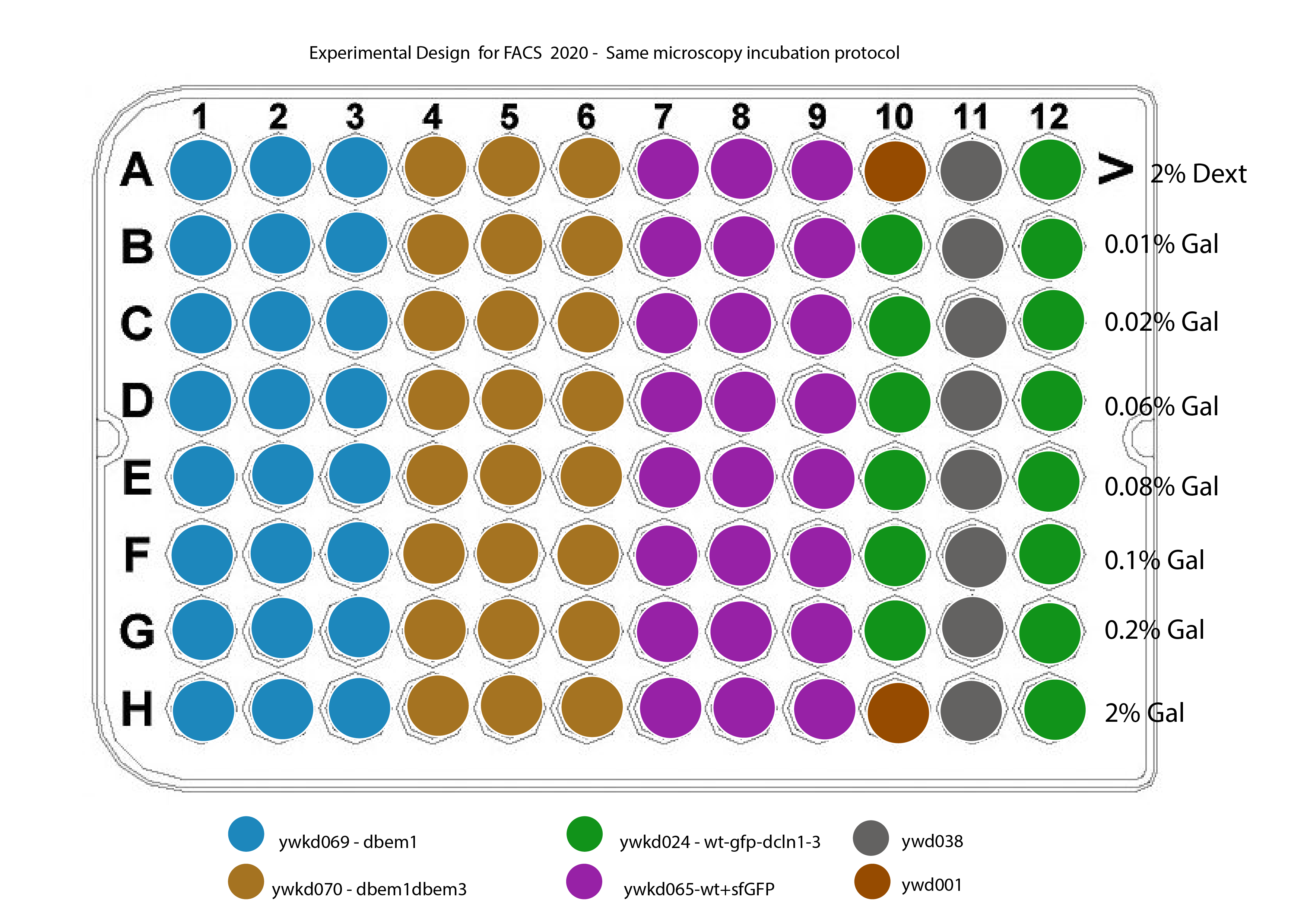 {#fig:plate-design width=50% height=50% }
{#fig:plate-design width=50% height=50% }
The strain ywkd024 will be measured to still compared with previous measurements done by Marit, in 2017.
Strains :
ywkd024 : RWS119 Wedlich-Söldner Lab collection a W303 can 1 1-100 his3 11,15 Galpr-myc-GFP-CDC42 YipLac204-MET-CLN2 cln1\(\Delta\)::HisG, cln2\(\Delta\), cln3\(\Delta\)::HisG (strain to compare with ywkd065(sfGFP))
ywkd065a New YWKD055c W303 URA-Gal1pr-sfGFP-Cdc42 sandwich (pWKD011 integrated) leu2 3,112 his 3 11,15
ywkd069 : New YWKD055c a W303 bem1\(\Delta\)::KanmX URA-Gal1pr-sfGFP-Cdc42 sandwich (pWKD011 integrated) MFAprHIS3 3,112 11,15
ywkd070 : YWKD070a,b,c New YWKD055c a W303 bem1\(\Delta\)::KanmX bem3\(\Delta\)::clonNAT URA-Gal1pr-sfGFP-Cdc42 sandwich (pWKD011 integrated) MFAprHIS3 3,112 11,15
ywkd038: RWS1421 Wedlich-Söldner Lab collection a W303 can1 1-100 his3 11,15 CDC42pr-myc-GFP-CDC42 YipLac204-MET-CLN2 cln1\(\Delta\)::HisG, cln2\(\Delta\), cln3\(\Delta\)::HisG (Reference for the native CDC42 expression)
Settings of the FACs experiment
Equipment-Model: BDFACSCelesta
Lasers: Alexa Fluor 488 at 495V
Flow Rate: 2ul/sec
Sample volume: 130ul
Plate: 96 well plate with flat bottom
# of events per well: 10000
FSC threshold:20000
FSC voltage: 407V
SSC volatge: 275V
Mixing volume: 65ul
Mixing speed: 200ul/sec
Nr. of mixes: 5
Actual procedure
1st Incubation , at 14:15 in 21012020 in 2% Gal.
Notes after this 1st incubation:
Neither ywkd001, ywkd038 and ywkd024 had enough cells after 24 hours of incubation in 2% Gal in 36C. (?)
2nd incubation, at 15:00 in 22012020 in the respective gal concentrations.
I washed two times the 1st incubation in CSM and CSM-met for ywd024 and ywd038 with 2% Raff, with no GAL.
I put the same amount of cells per strain in all conditions, aka I pipetted the same volume of a concentrated cell culture (from 5ml to ~400ul) in every tube of the same strain.
For ywkd024 I could not pipette any cell in 0.1% Gal and 0.2% Gal, because I ran out of cells :( but this is not a big problem anyways.
23012020: I diluted ywkd065 and ywkd070 50x at 10:30 , because they were a bit too dense for FACs. The rest of the strains were not dense at all.
at 12:30 Inoculate 200ul in each well of the 96 well plate according the layout (see @fig:plate-design).
60.4. Results#
60.4.1. Plate data#
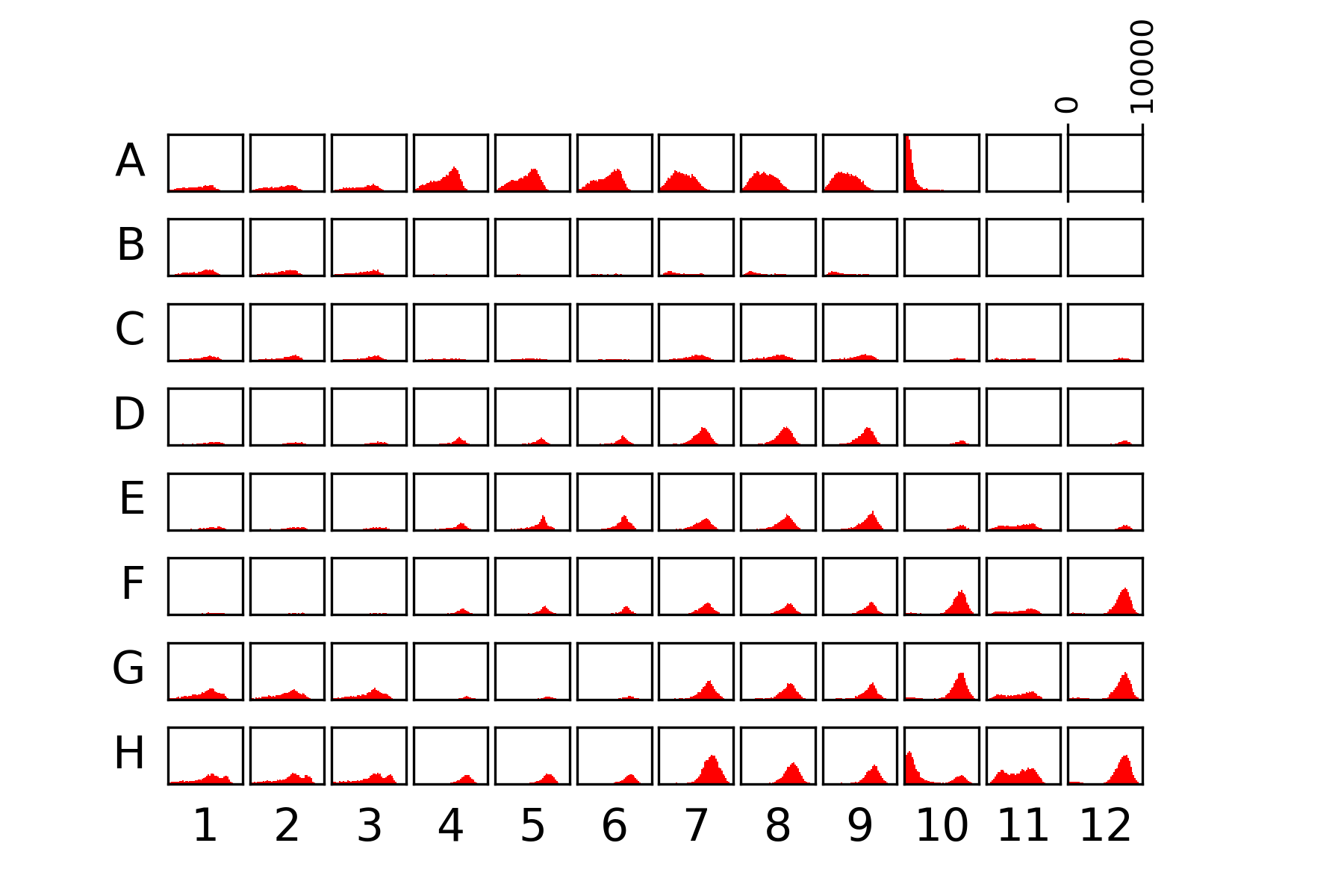 {#fig:whole-plate width=100% height=50% }
{#fig:whole-plate width=100% height=50% }
60.4.2. Box plots#
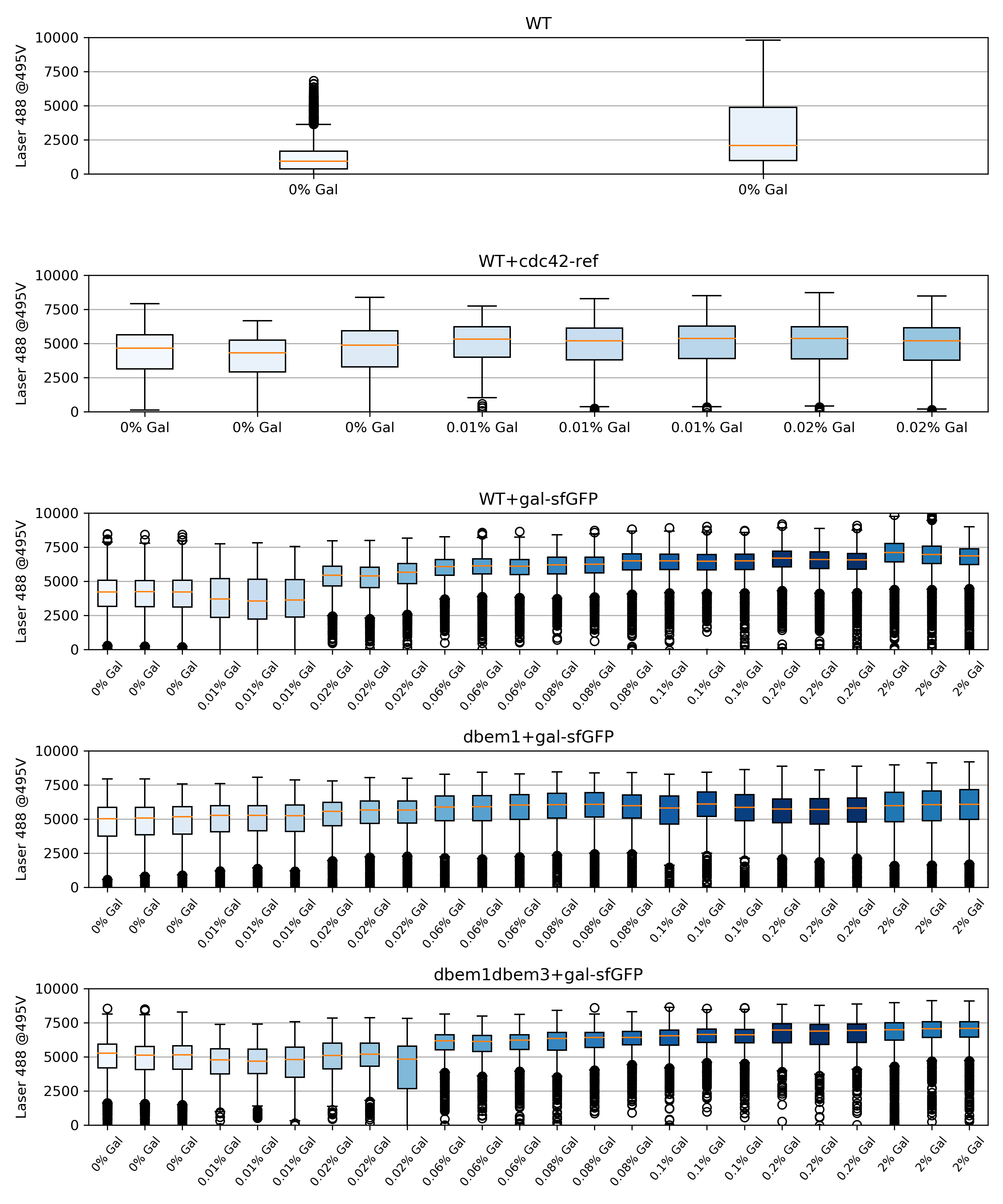 {#fig:box-plots width=100% height=100% }
{#fig:box-plots width=100% height=100% }
60.4.3. Histograms#
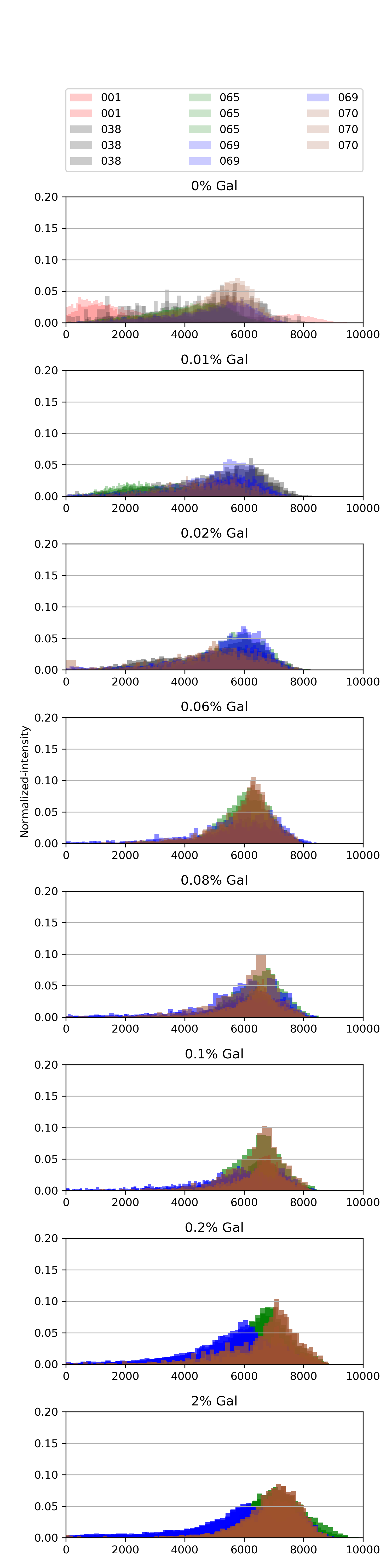 {#fig:histograms width=100% height=100% }
{#fig:histograms width=100% height=100% }
60.4.4. Medians per strain#
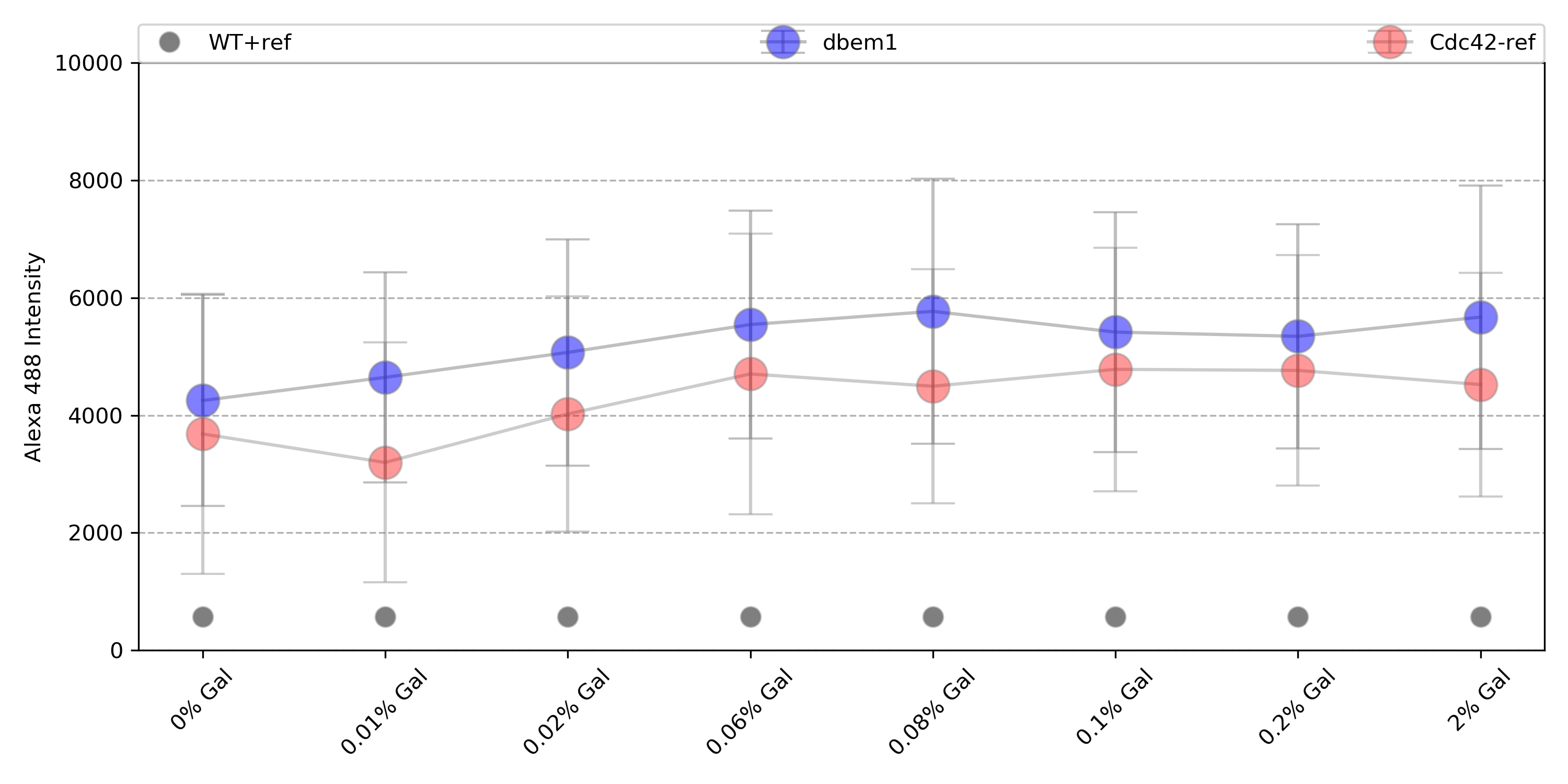 {#fig:dbem1}
{#fig:dbem1}
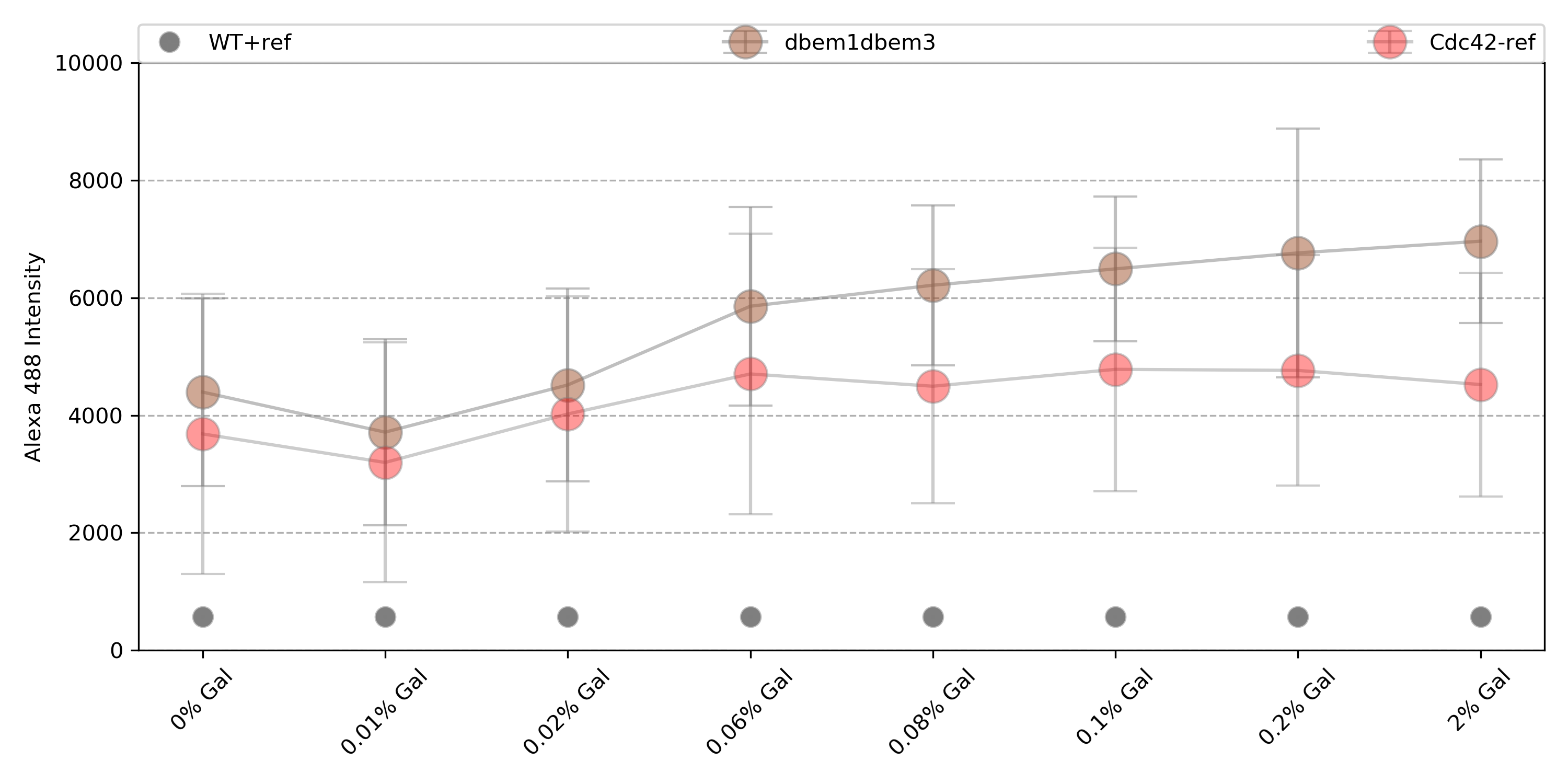 {#fig:dbem1dbem3}
{#fig:dbem1dbem3}
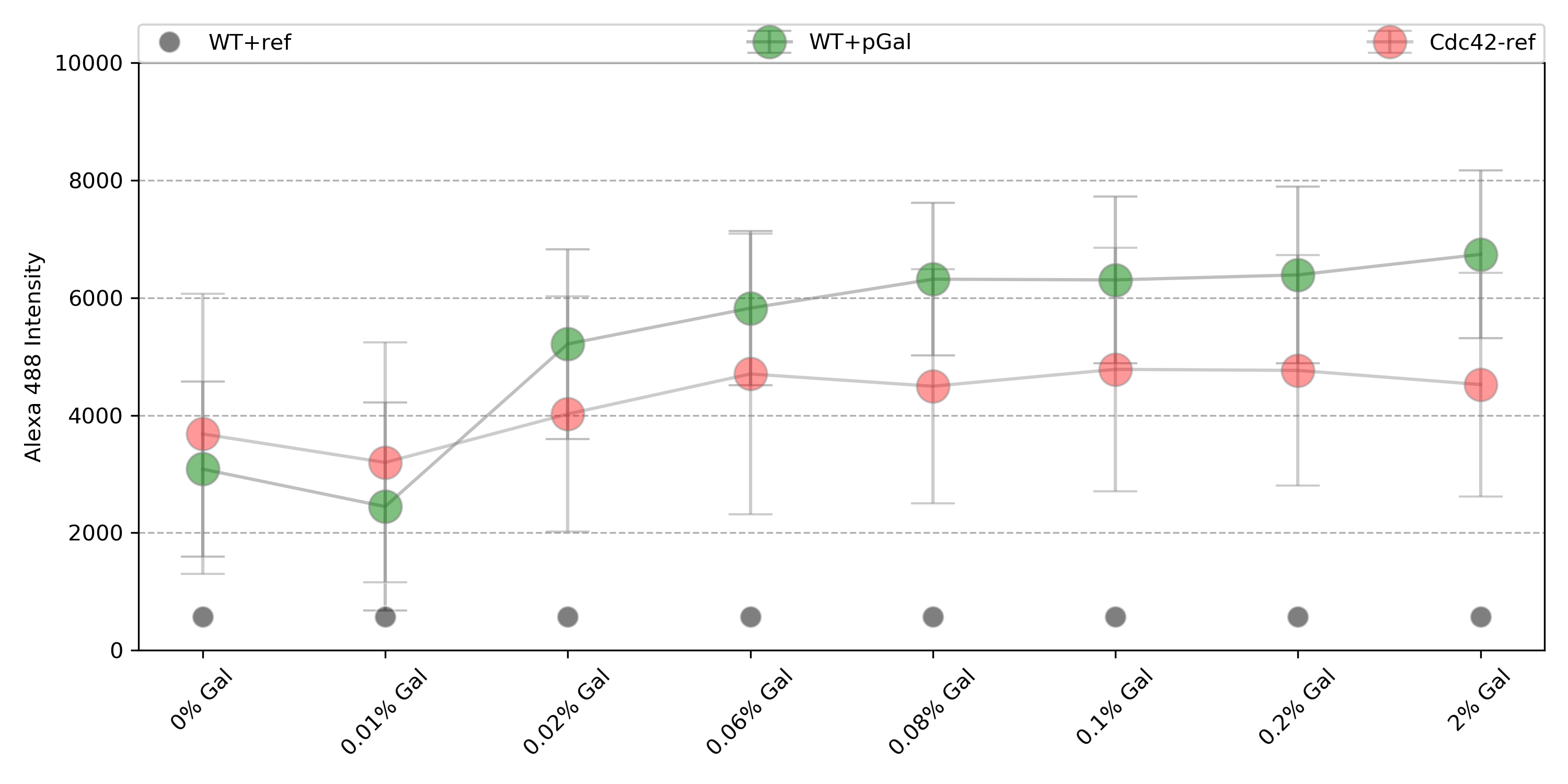 {#fig:wt_gal}
{#fig:wt_gal}
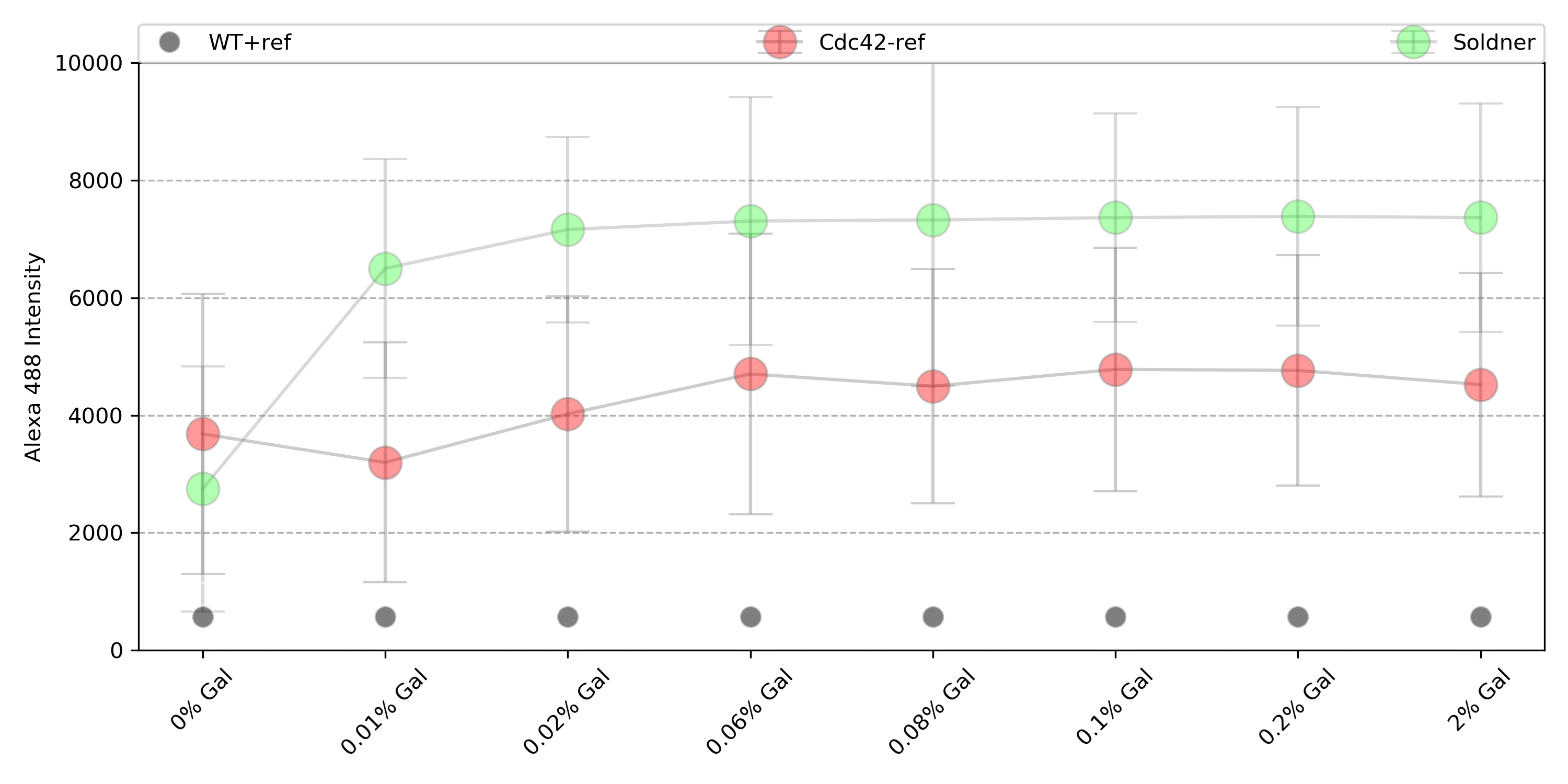 {#fig:024}
{#fig:024}
60.4.5. Cdc42 relative expression#
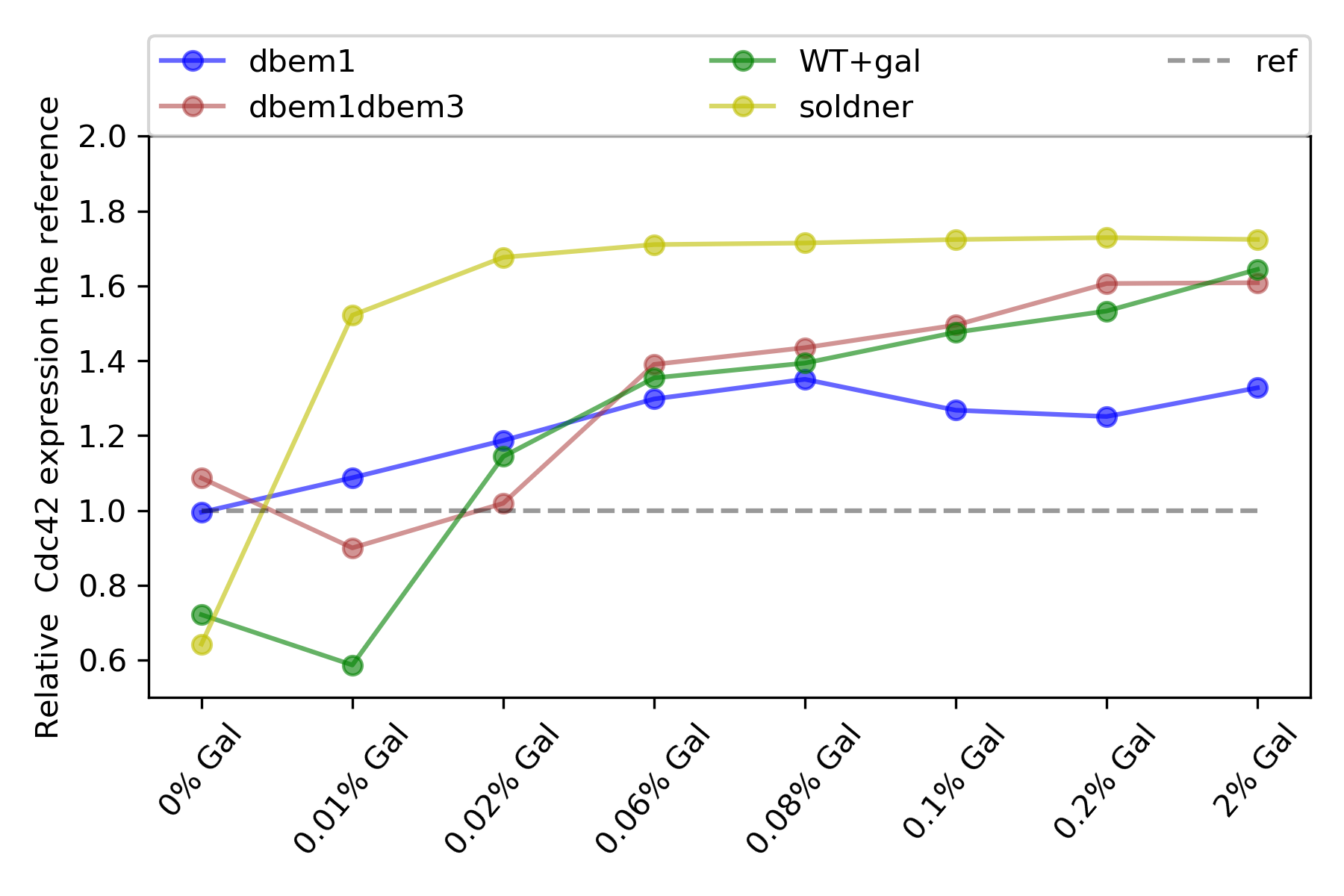 {#fig:cdc42-expression}
{#fig:cdc42-expression}
60.4.6. Total counts#
The maximum amount of counts, or cells that the FAcs was set to analyse was 10000, which corresponds with an OD=~ 0.1. Assuming that OD=1 is around 10\(^6\) cells in 1mL. So in 100ul , for OD=1, it has to have around 10\(^5\) cells. Hence 10000 (10\(^4\)) cells will be an OD=~0.1.
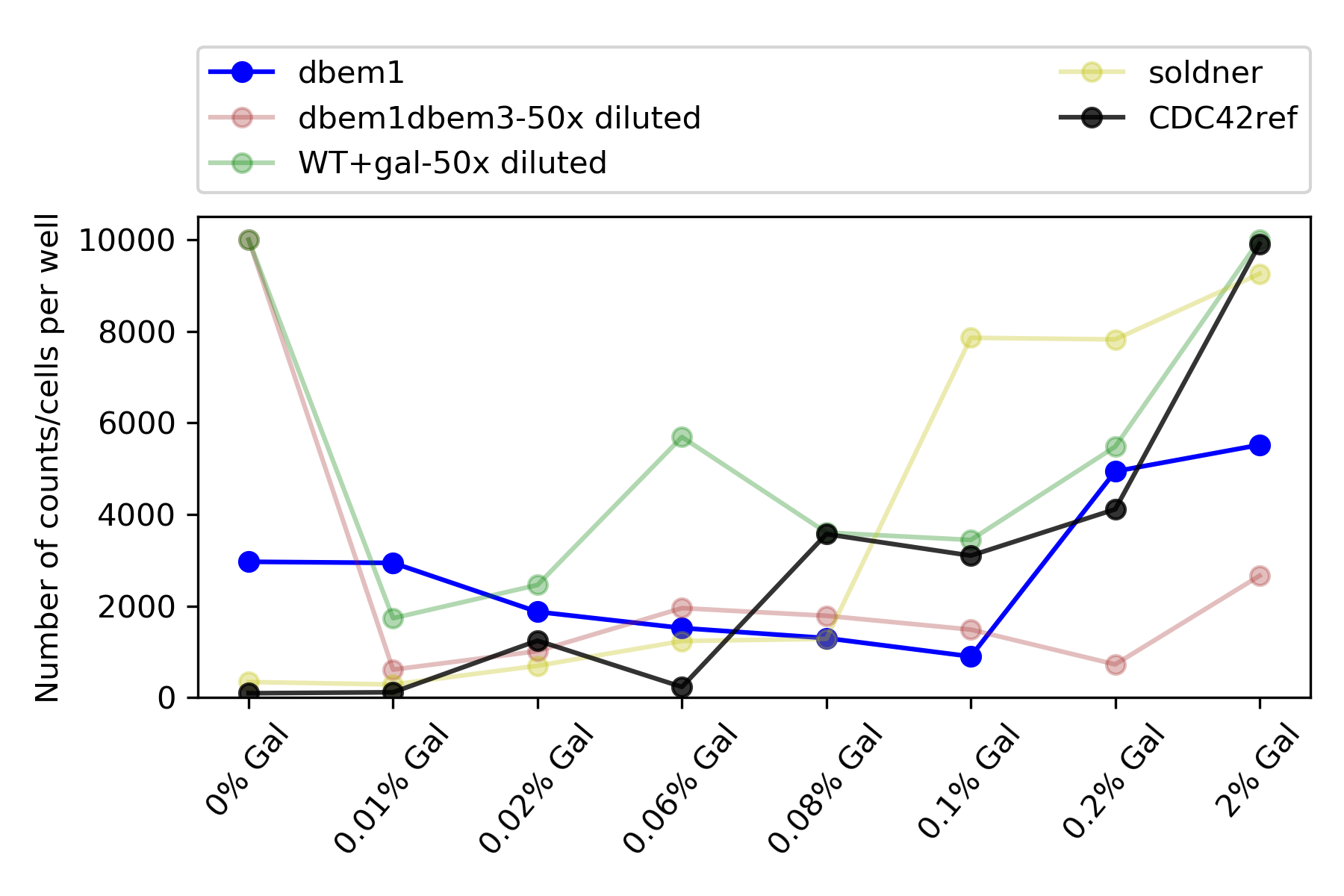 {#fig:counts}
{#fig:counts}
Only dbem1dbem3, WT+gal has reached the maximum of counts for 0% Gal, while dbem1 did not. This indicates that the leftover of Cdc42 from the previous 2% gal incubation was not enough for dbem1 to keep growing.
60.5. Conclusion#
Basically the dynamic range of the promoter did not change from previous experiments, no more than 2.5 fold change.
Now , it can be seen more difference between dbem1-dbem1dbem3-WT+gal in their expression.
dbem1+Gal did not change too much the cdc42 expresion across galactose concentrations, as the other do.
Interestingly the dynamic range, from 0% gal to 2% Gal, in average increases from the dbem1 to the WT+GAl, as follows:
dbem1: 1.29 fold change
dbem1dbem3: 1.58 fold change
WT+gal: 2.18 fold change
Soldner strain: 2.64 fold change
ywkd038-cdc42 reference: 1.22 fold change (should be 1)
For ywkd065:
Gal concentration |
[cdc42-gal]/mean-ref |
|---|---|
0% Gal |
0.541095 |
0.01% Gal |
1.310883 |
0.02% Gal |
1.391903 |
0.06% Gal |
1.553951 |
0.08% Gal |
1.618289 |
0.1% Gal |
1.624045 |
0.2% Gal |
1.572428 |
2% Gal |
1.469435 |
