Title : gDNA extraction of yll138
Contents
29. Title : gDNA extraction of yll138#
29.1. Date#
17112020
29.2. Objective#
-To have gDNA to make the PCR in order to transform ylic133 and make a bem3\(\Delta\) for SATAY.
29.3. Method#
Genomic DNA extraction using the Roboklon KIT
I pulled the pellet of 4ml of culture for one extraction.
I used in total 4 tubes with 4ml of culture
I preheated the elution buffer at 55C before eluting it.
I eluted 10ul per tube , so in total I have around 40uL of DNA.
Nanodrop concentration: 15ng/uL
A260: 0.3
A280: 0.1
260/280 : 2.6
260/230: 1.6
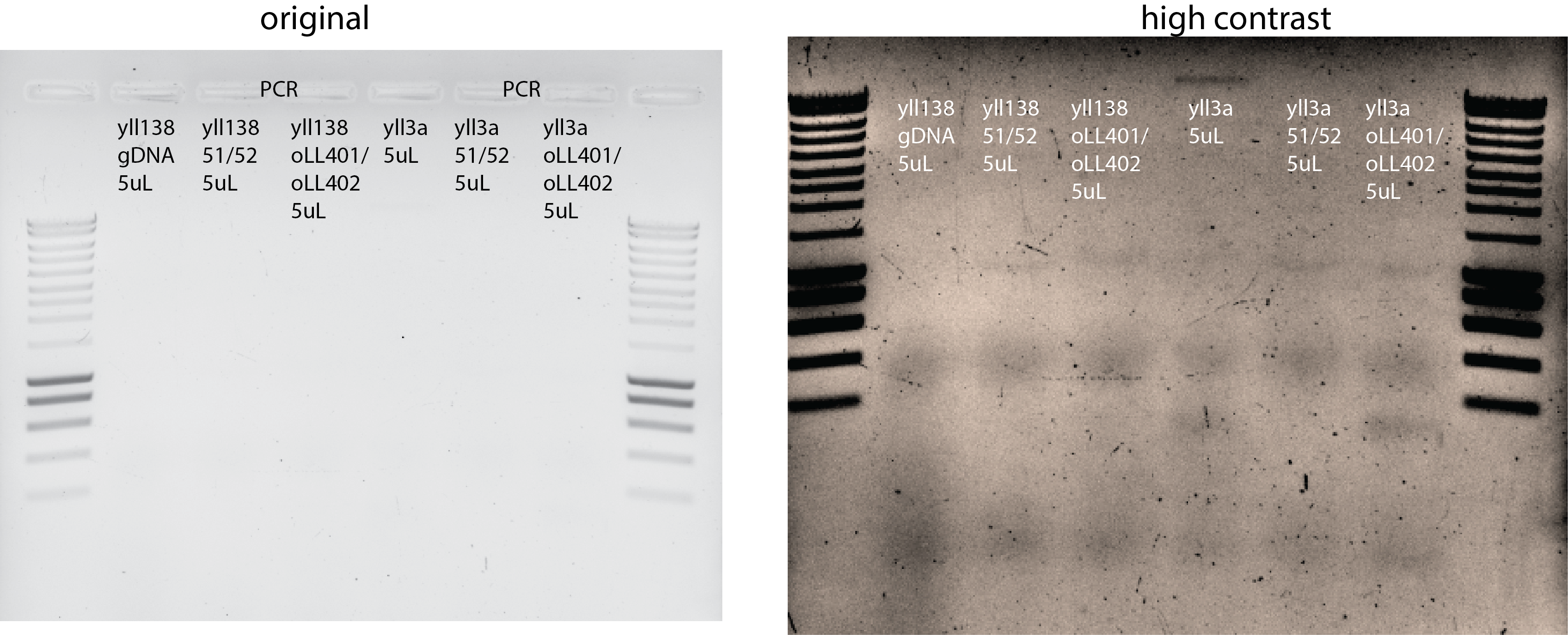
PCR
Protocol Leila (block A machine upstairs)
95C 3mins
do 35x cycles:
95C 30secs
55C 30secs
72C 1min
end cycle
72C 5mins
12C on hold
2ul of DNA
Primers 51/52 and oLL401/oLL402
Adding also 2uL of DNA from yll3a as control.
29.4. Results#
29.5. Next steps#
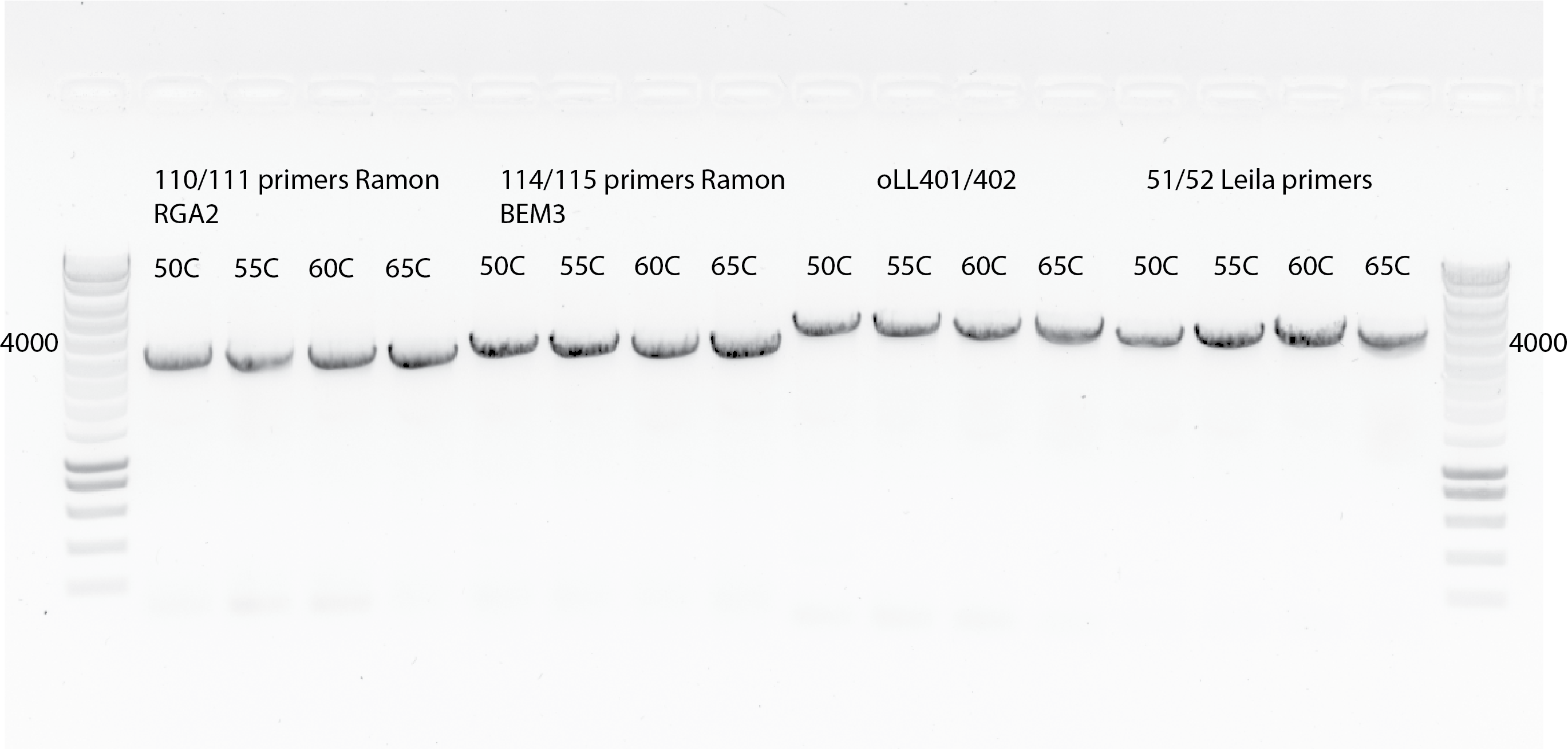
Annealing temperature gradient , with the following T: 50C, 55C , 60C and 65C with 2 mins of extension time. Before was 1 min.
Also for the PCR , we used a new dilution of all primers (always vortexing them before diluting.)
Using Ramon primers(110/111,114/115), Liedewij primers(oLL401/oLL402) and my primers (51/52).
New genomic DNA extraction (23112020)
1.5ml of a highly dense culture per eppi
5 DNA extraction per strain (yll138 and yll3a)
30ul of elution
concentration on gel (5ul loaded) using BioRad
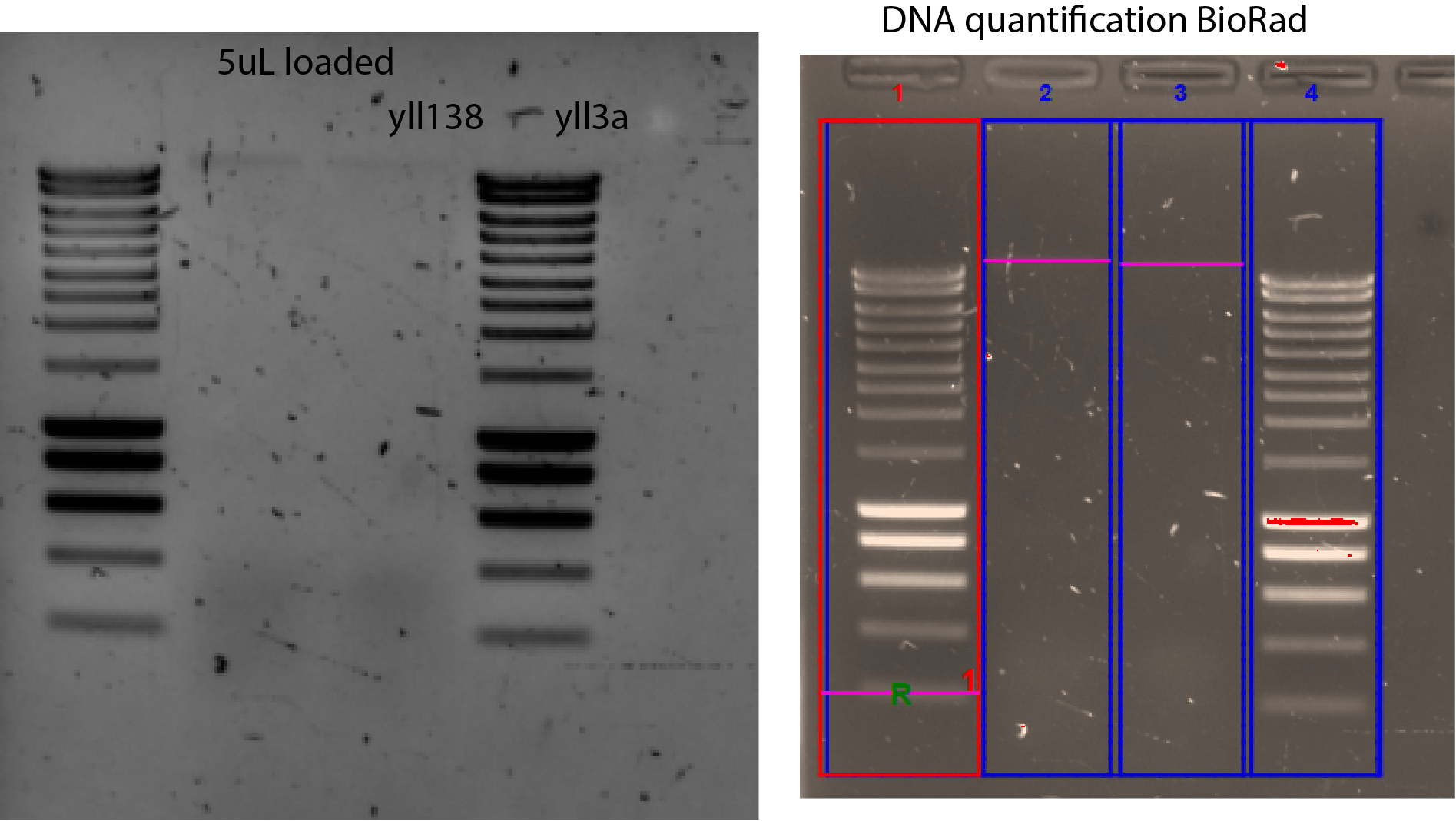
yll138 intensity = 0.4 x Intensity (20ng band) = 8ng/5ul ~ 1.5 ng/ul
yll3a intensity = 0.2 x Intensity (20ng band) = 4ng/5ul ~ 1 ng/ul
These are really low numbers
PCR with those genomic DNA and the old DNA that previously worked!
Use 5ul DNA template for the new extraction and 1ul of the old yll3a stock
Use oLL401/oLL402 and 51/52 primers , newly diluted from the original stock.
Use 60C annealing temperature and 2 mins in 72C for the extension.
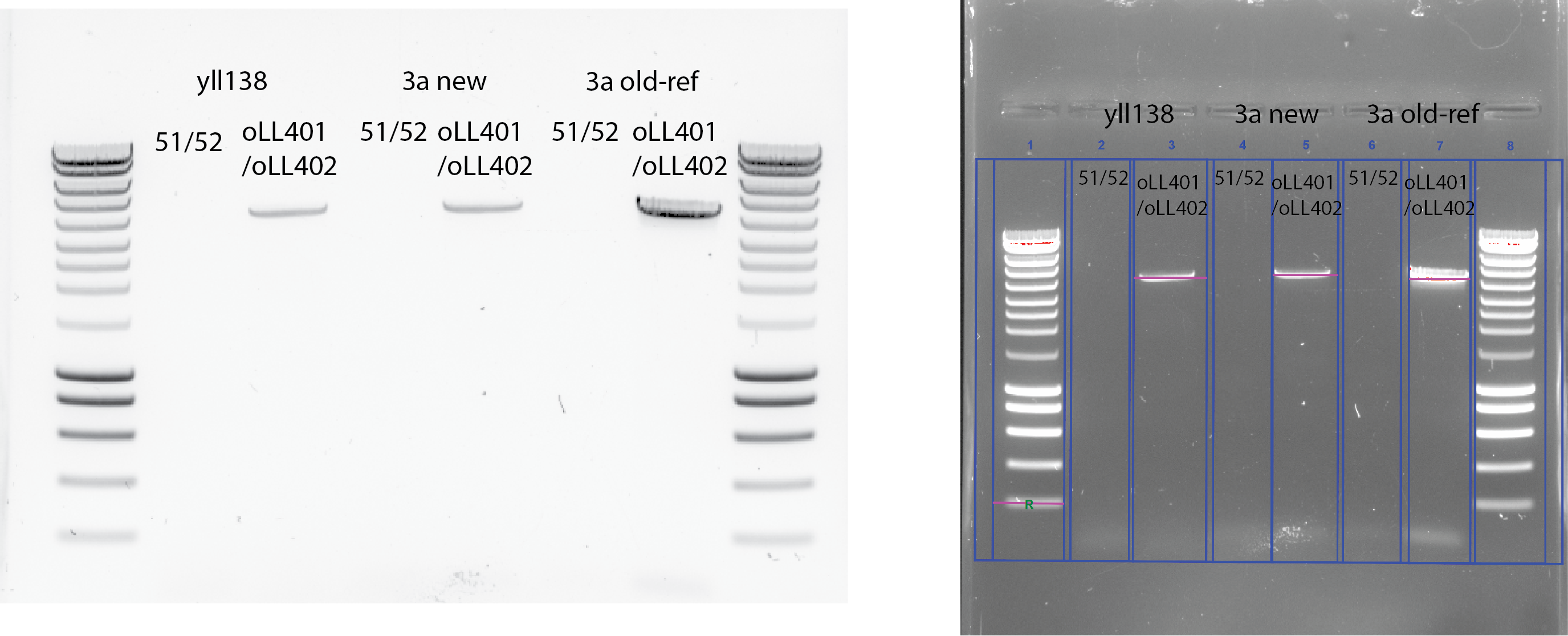
My primers 51 and 52 did not work while Liedewij’s primers did work.
Concentration of the PCR product using BioRad
Intensity(yll138)= 12.66 * Intensity (20ng)=253.2/5ul=50.64 ng/uL
Intensity(yll3a)= 13.43 * Intensity (20ng)=268.6/5ul=53.72 ng/uL
One interesting point from this result is that the length of the PCR product from yll138 that have bem3::NAT is the same as the WT. From previous PCR the PCR product size with 51/52 primers is roughly the same as with oLL401/oLL402 in the yll3a (old reference) template.
If I have enough PCR product to do both transformations I will continue with it , and check if they acquire NAT resistance , then I will know they are the right clones. Anyways I will send to sequence those clones and I will see what is in there.
29.5.1. Troubleshooting the PCR#
Another PCR with 10ul and 1ul from yll138 DNA and 10ul and 1ul from yll3a DNA (old reference) with primers 51 and 52.
To test more template and less buffer with the 1ul DNA that may hinder the efficiency of the PCR using yll138 as template.
Repeat 8 times the PCR using yll138 as template (5ul) with oLL401/oLL402 to have more PCR product for the transformation.

29.6. Next Steps#
Use a different strain for transformation , this one is causing too much problem and also not giving the right product as it has the same length as the WT, which should not be , given that BEM3 is not there. I will use the one from Wessel that has worked.
29.7. Conclusion#
It seems the extension time placed a role in the PCRs
I wont continue with this strain (yll138) for transformation.
I was overlooking the first step of the genomic extraction kit , which is the activation of the column with Buffer BG. See here the protocol for more details. So this was the reason why I was having so low yield. 😣😣