Title: Checking bem2 deletion on the transformed strain ylic133_5
Contents
45. Title: Checking bem2 deletion on the transformed strain ylic133_5#
45.1. Date#
30042020
45.2. Method#
Colony PCR with specific primers from inside the leu2 marker and the bem2 gene. Check the documentation for the transformation
This is now possible because I have sequenced the transformed strains that grew in -leu2 , and I have the right sequence to design the correct primers. Previously we couldn do it because the leu2 marker was taken from a plasmid but the marker was not completely annotated where it starts and ends.
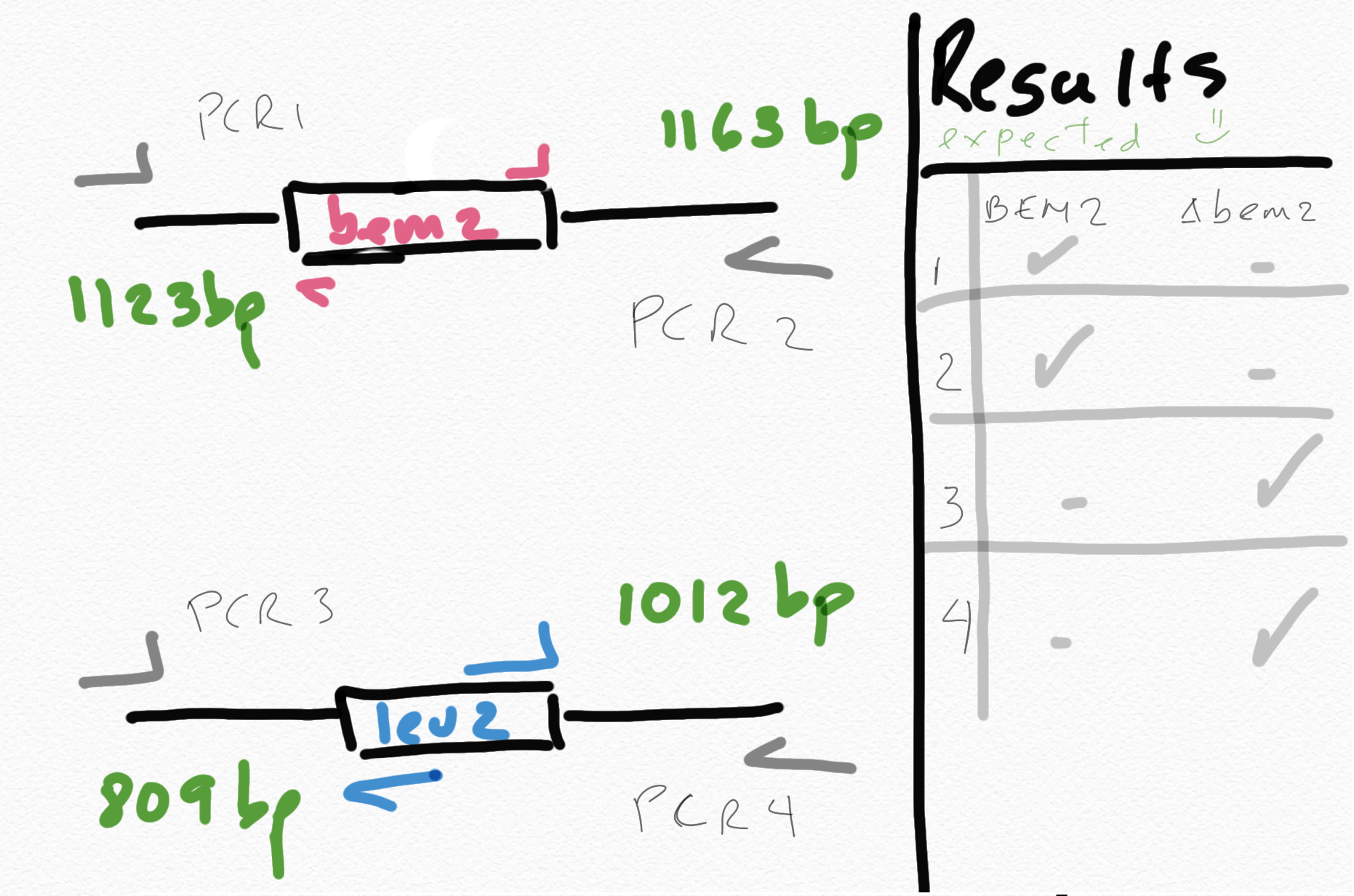 {#fig:sketch-pcr}
{#fig:sketch-pcr}
45.2.1. Procedure#
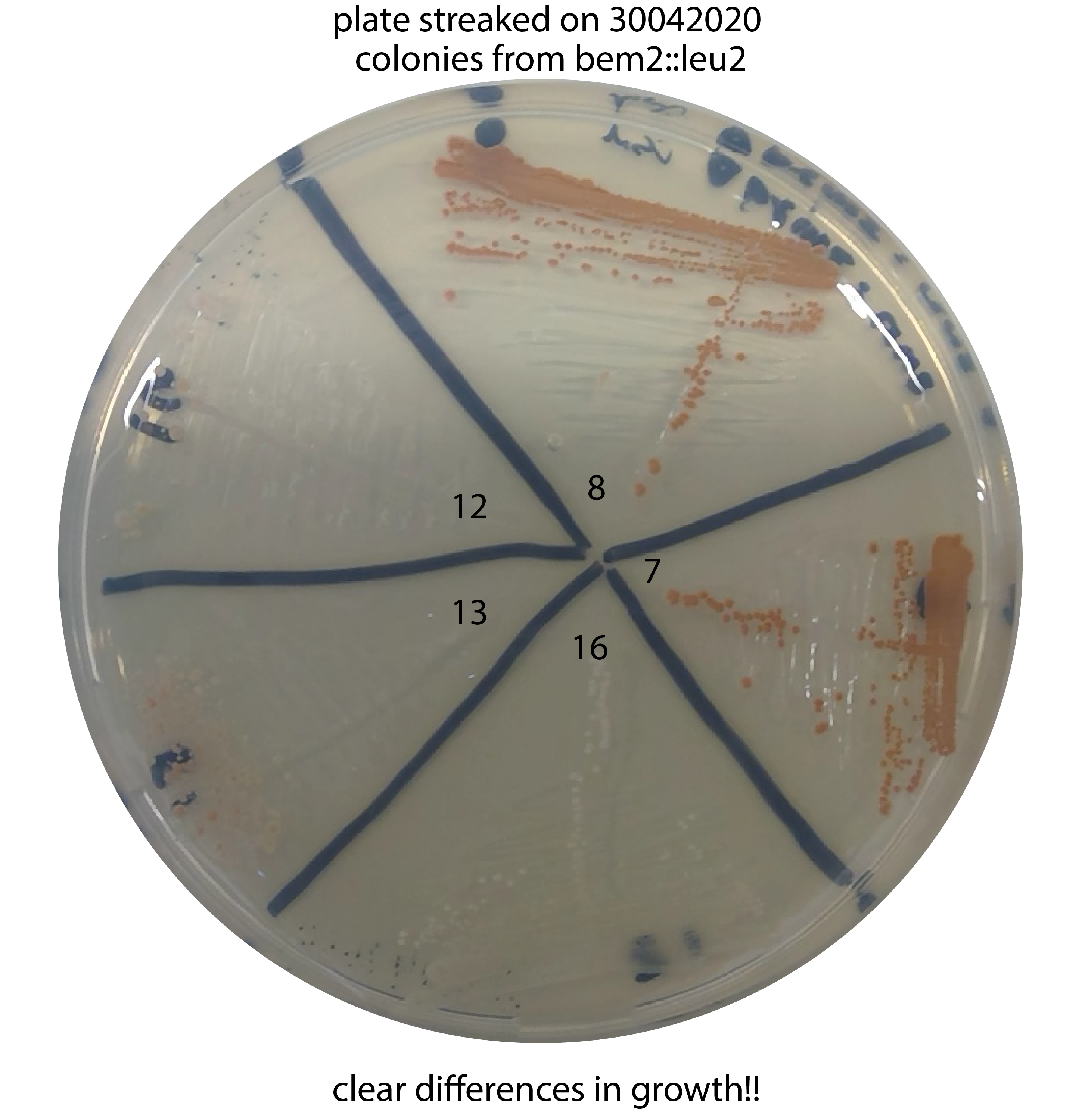
30042020 - Streak out the glycerol stocks of all the colonies from ylic135 (colony 7,8,12,13 and 16) on CSM-leu2 plate and also ylic133 (WT ade2-) in YPD for further colony picking for the colony PCR.
04052020 - Result of the growth in plate:
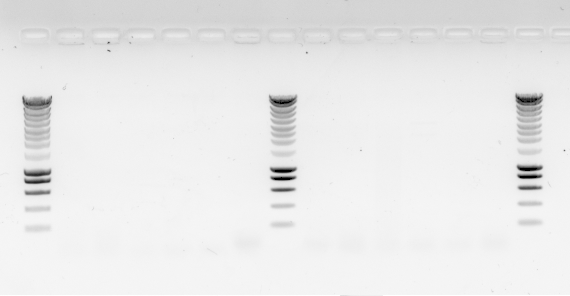
04052020: Colony PCR
12 PCRs in total because I have 6 samples for every set of primers. One positive set using 5 colonies from bem2:leu2 and ylic133 and primers 49-50 for leu2 check. And a negative set with the same samples but woth primers 47-48 for BEM2 check.
Protocol : Leila (name in the machine)
No bands in the PCR gel (?????)