Title : Genetic checks for a single colony from the stock of bem1:KanMx mutants
Contents
10. Title : Genetic checks for a single colony from the stock of bem1:KanMx mutants#
10.1. Date#
29102021-
10.2. Objective#
To figure it out if the stok completely is unstable , that is , all the cells are aneuploidy after bem1:kanmx transformation.
If not , then store a new clean glycerol stock.
10.3. Method#
Plate from glycerol stocks in CSM+2%Raff+2%Gal+G418
Plates were tested with negative(yll3a) and positive control (ywkd017)
Take one single colony and inoculate in same liquid media over the weekend
gDNA extraction (ylic139b,ylic140a,ylic140b,yTW001a)
2 eppies per strain
30ul elution
42-47 ng/uL DNA concentrations
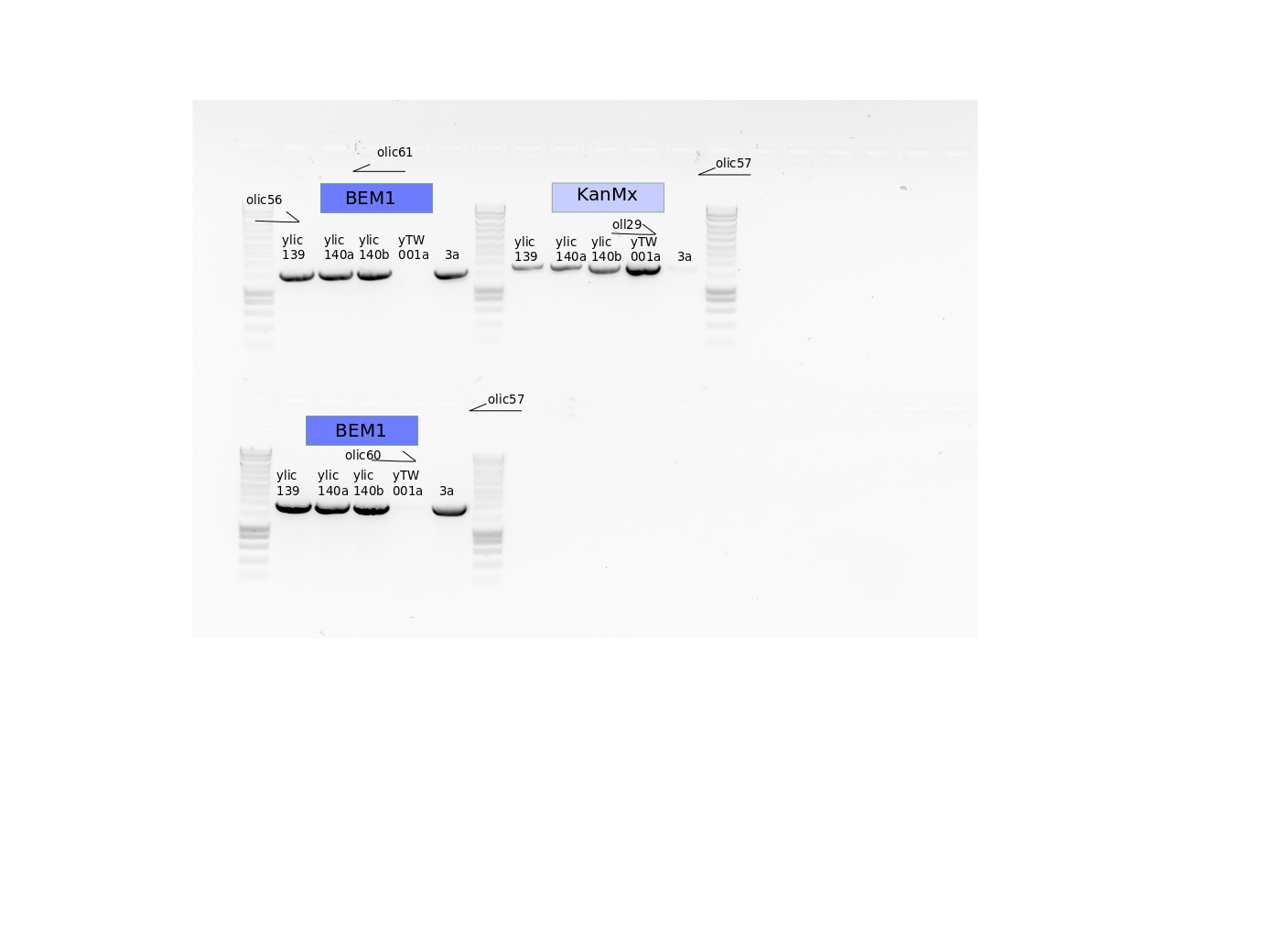
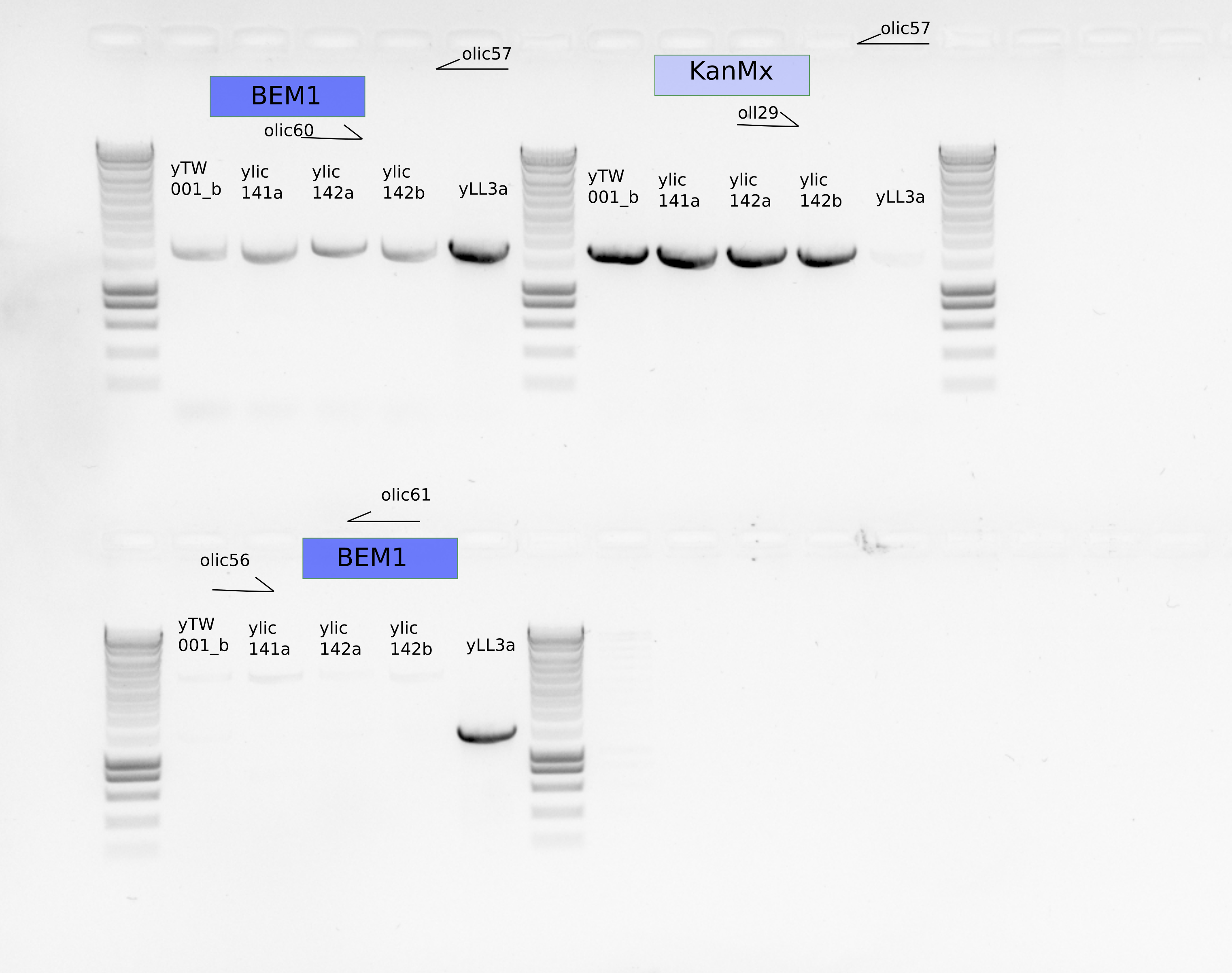
PCR inside BEM1 and inside KanMx
olic56/61, olic57/60, oll29/olic57
1ul DNA
glycerol stocks if OK
Only for yTW001a
Day 2:
Inoculate a 2nd colony from ylic139a(discarded)
Analyze ylic141a,b and ylic142a,b
10.4. Results#
10.4.1. Sequencing#
PCR to prepare samples for sequencing:
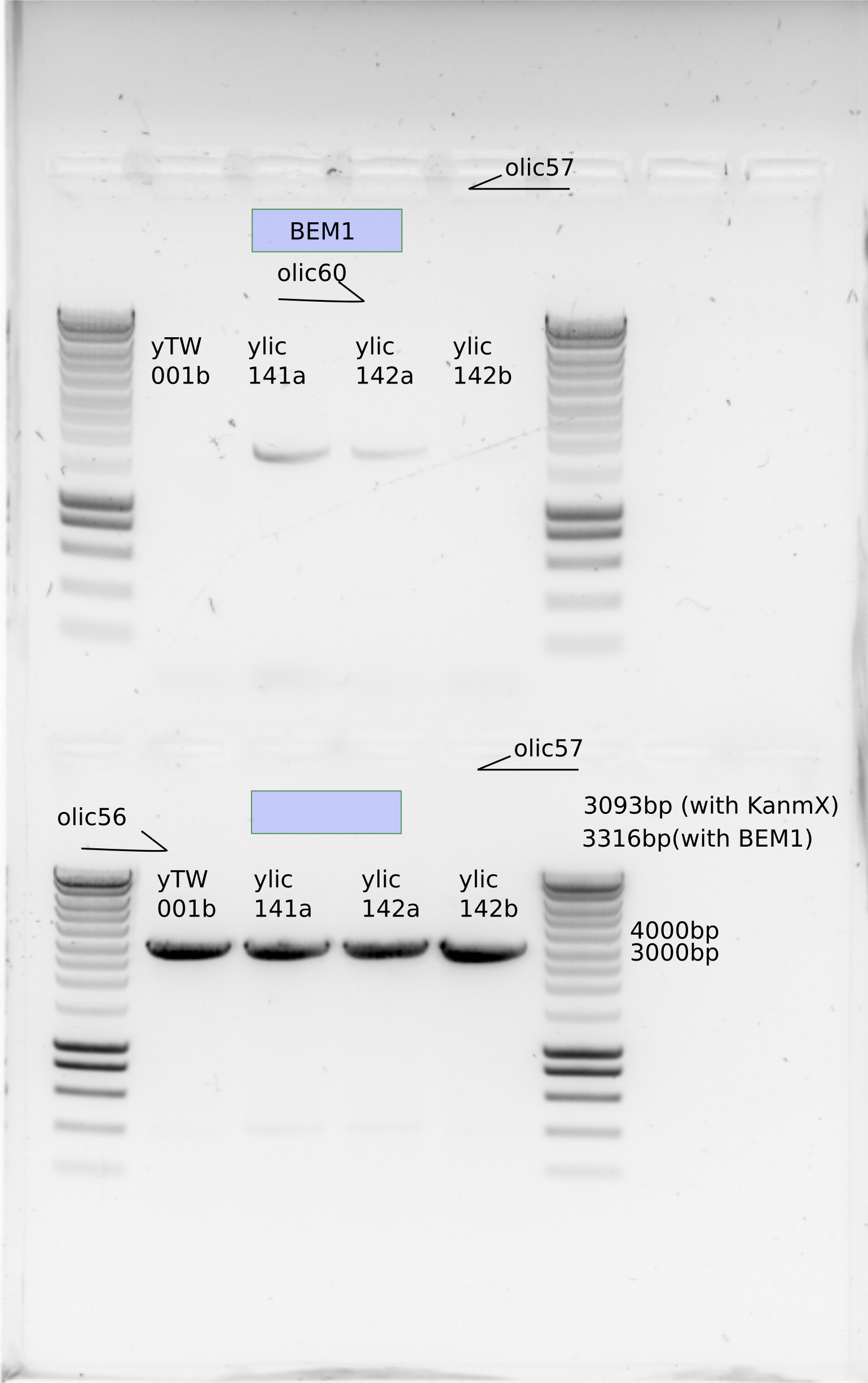
yTW001b seems that do not have BEM1
I sent samples from ylic141a and ylic142a with olic60 and olic57 primers to sequencing.
I sent all the samples from PCR from olic56 and olic57 to sequencing.
This is indicating that both strains ylic141 and ylic142 are aneuploidies, because they show the presence of PTEF from KanMx and the downstream region of BEM1 in the same strain.
10.5. Conclusion#
The pgal mutants are unstable clones, yet, yTW001a seems to be rescued.
To confirm that BEM1 is not in the SATAy mutants , I will sequence using olic60/olic57 and olic56/olic57 primers.
yWT001b seems that do not have BEM1.
ylic141 and ylic142 are aneuploidies.
Repeat the transformations fro ylic141 and ylic142 ,and check for every colony the presence of Kanmx and the abscence of BEM1.
