Title: 26062019-ylic132 transformation with the OEP2 (unsuccesful) :x: :disappointed:
Contents
91. Title: 26062019-ylic132 transformation with the OEP2 (unsuccesful) :x: :disappointed:#
91.1. Date#
26062019
91.2. Objective#
To remove the ura3 gene from ylic132= ade2- ura3+ by inserting an empty sequence in the ade2 locus , where the ura3 gene is in ylic132.
91.3. Method#
Yeast transformation protocol
I used in this case 150ml of overnight culture.
91.3.1. Homology arms:#
left homology arm: 252 bp
PCR of primer_1_new_upstream_forward and primer_7_upstream
right homology arm: 271 bp
PCR of primer_6_new_downstream_reverse_NO_Rga1 and primer_8_downstream
91.3.2. DNA concentration#
4 ug
91.3.3. starting OD#
OD=1.8
91.3.4. Selection plates#
-URA + 2x ADE (for ylic132 negative control- no transformed cells )
-URA + 2x ADE for a positive control using yll3a with a URA plasmid I received from Ramon.
5FOA plates + 2x ADE , in which only cells lacking URA3 are capable of grow. (positive control for the transformed cells)
91.4. Results#
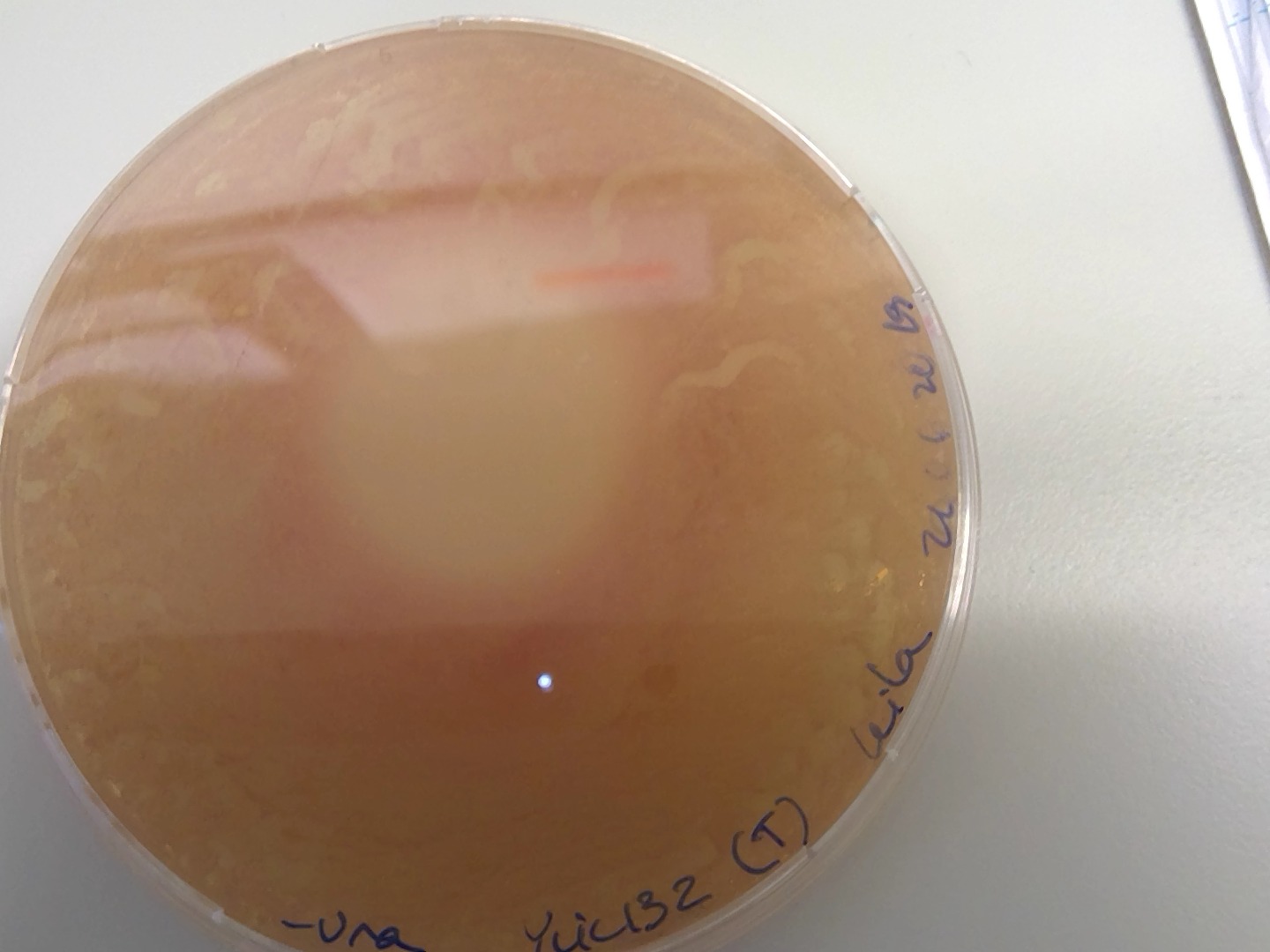
One single colony in 5FOA plates for the transformation plate , of ylic132T ☹️
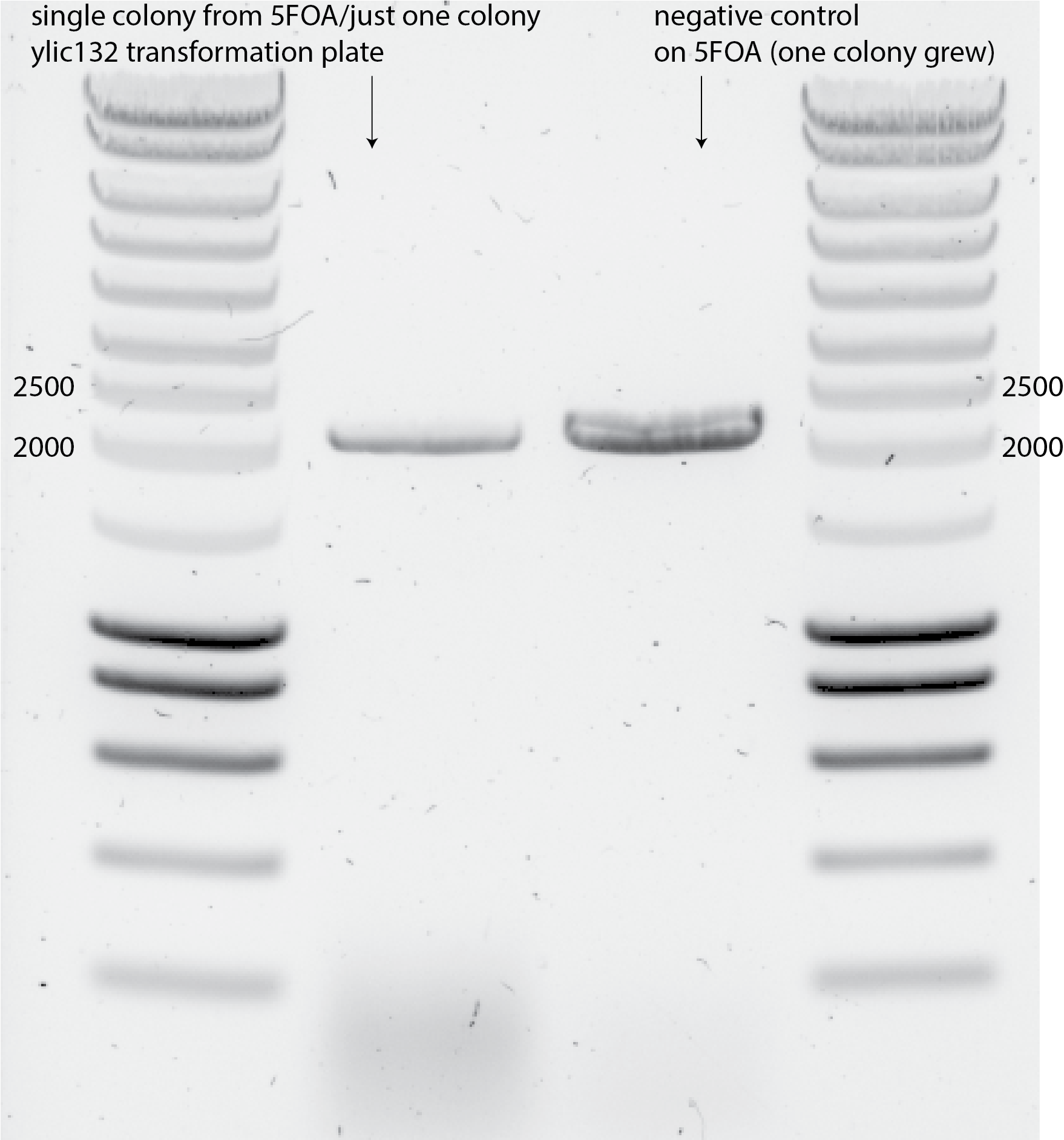
Colony PCR with 5ul of the single colony in 50ul MiliQ. Using primer_9_ADE_forward_check and primer_10_ADE_reverse_check, the PCR product length if URA3+promoter is still in the genome is: 2065bp. This is the length is observed when running that PCR on the transformed single colony and the single colony for the negative control that grew on 5FOA plate
One single colony in 5FOA plates for the negative control plate 5FOA with non transformed cells -> ura point mutation
No colonies neither on the positive control plate, with yll3a + ura plasmid given by Ramon (☹️) he says the plasmid extraction yields very low concentration though
For the -ura plates I keep seeing the fried egg like structure 😒with a lighter spot on the plate and pink cells around.
Explanation: I did not do the recovery step required when transforming with a dominant marker. The cells before plating need to be in YPD for at least 2 hours and then plating on the selection plate, in this case 5FOA. Look here
91.5. Next Steps:#
Redo the transformation without skipping the recovery step of growing the cells in YPD before plating them .
Do 2 transformations using both recover steps:
Growth 200ul cells in 1ml of YPD during 1-4 hours, in 30C, then spin down the cells and resuspend in sterile MiliQ and then plate them using 20ul cells+150ul MiliQ, and then 180ul cells
Resuspend the cells in 1ml of YPD. Plate 200ul culture in YPD and next morning replica plate in the selection plate. And save the rest of the cells in the 1ml YPD for overnight culture and plating next morning in the selection plate.