Title : 24092019- pBK549 transformation on ylic133 for further sanity check in SATAY. :pensive:
Contents
67. Title : 24092019- pBK549 transformation on ylic133 for further sanity check in SATAY. :pensive:#
67.1. Date#
24092019-30092019
67.2. Objective#
To ensure that the constructed strain is able to pass the Satay sanity check , and then I can continue with the further steps, like mating with yEK7a.
67.3. Method#
24092019 16:30 Plating in YPD plates all glycerol stocks of the different biologcal replicates.
26092019-Results of the plating in YPD
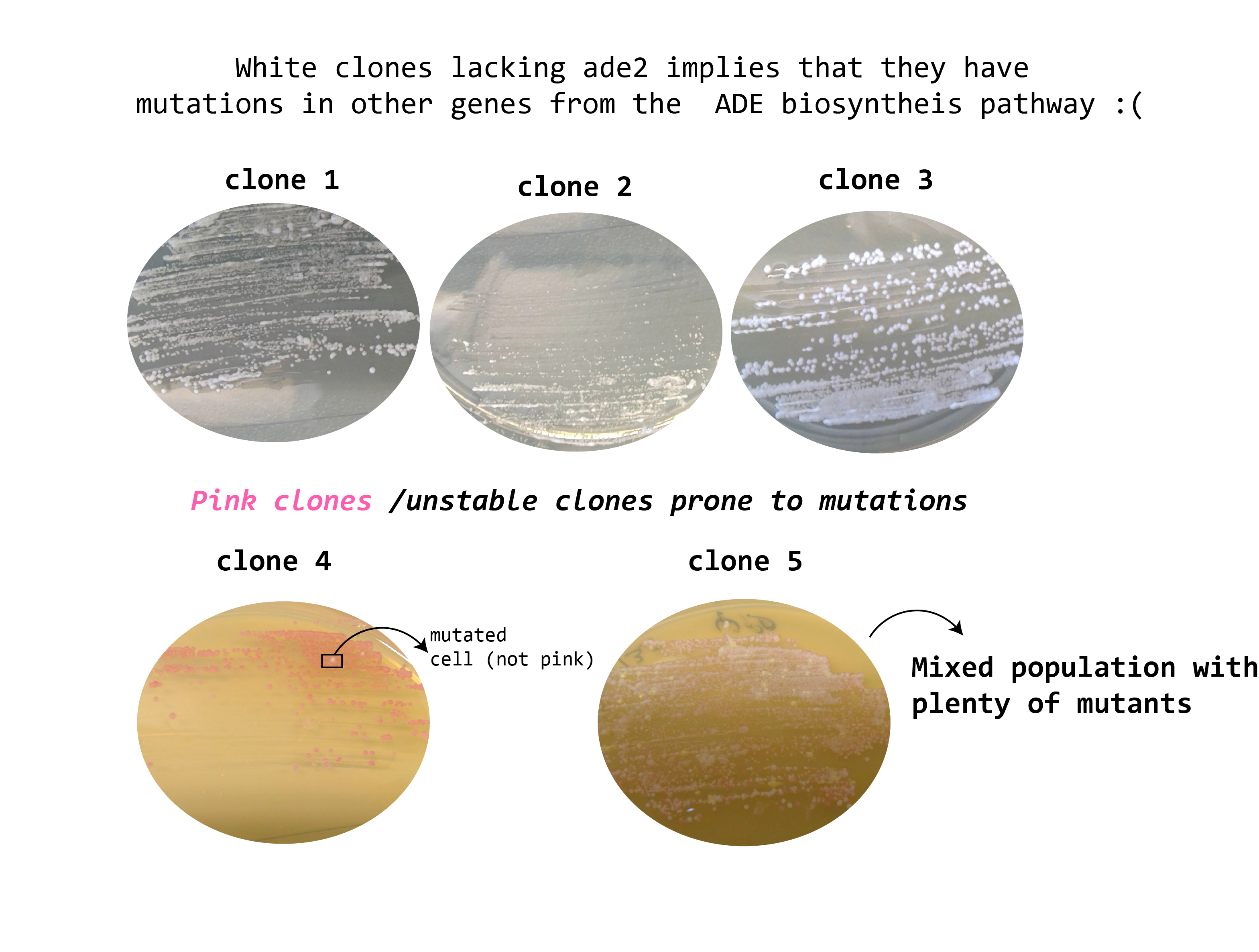
26092019
13:50 Inoculation of one single colonie from ylic133_4 plate, and a bit more than a colony of ylic133_4 as well , Byk832 and one single white colony from ylci133_1 in YPD+6xADE (that was the amount Enzo used for the last succesful pBK549 transformation=0.055g of ADE in 500 ml of media) Look here into the pBK549 transformation on 05082019. This is to decrease the changes of supressors that supress the pink phenotype. This inoculation is to transform them with pBK549 plasmid.
Use Byk832 as a positive control for the transformation. Inoculate it at the same time as the pink ylic133 clones and one yellow to reconfirm that this clone can not be used for SATAY.
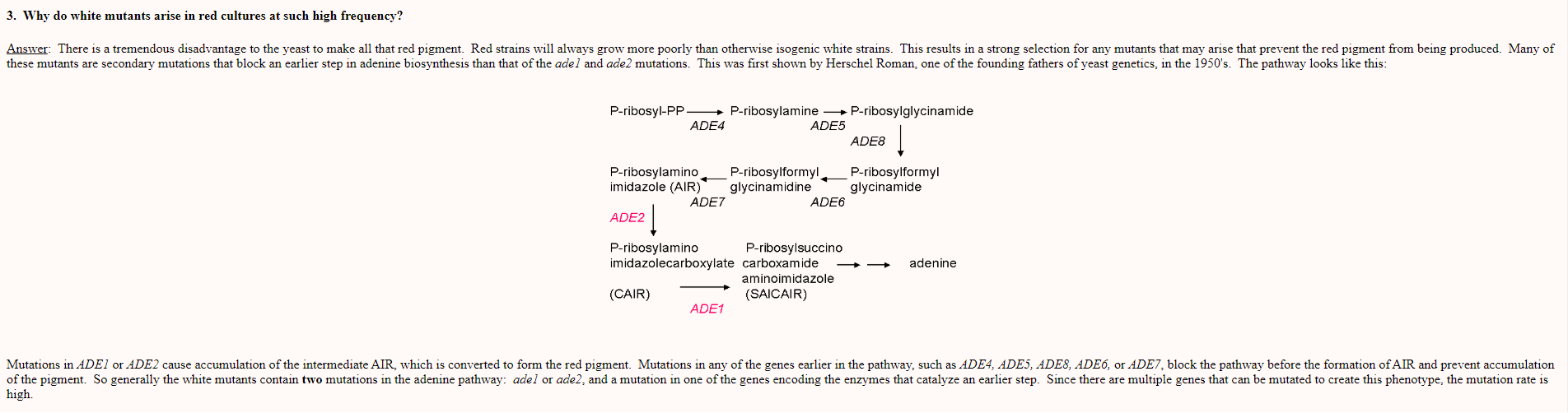
See the following picture from this page to confirm that the yellow phenotype I see in some of the ylic133 clones is because mutations in some other ade genes in the adenine biosynthesis pathway.
26092019 - restreaking of different colonies from clone 5 of ylic133, to hopefully clean up the glycerol stock.

30092019 store one colony from the pink restreaking as new ylic133_5 glycerol stock
27092019 - pBK549 plasmid transformation
OD measurements 9:55
OD-10X dilution
Titer
Dilution factor to OD=0.5
Time
ylic133_1
1.263
12.6
25.2
9:55
ylic133_4 (1)
1.843
18.4
36.8
9:55
ylic133_4 (2)
1.417
14.1
28.2
9:55
Byk832
0.063
0.6
1.2
9:55
The ylic133_4 tubes were not pink yet, I think this make sense due to the excess of adenine that the media have.
10:15 1st incubation until around 14:15 , to reach log phase for transformation.
13:30 OD measurements
OD-10X dilution
Titer
Time
ylic133_1
0.158
1.6
13:30
ylic133_4 (1)
0.088
0.9
13:30
ylic133_4 (2)
0.140
1.4
13:30
Byk832
0.063
0.34
13:30
I diluted 2x Byk832 down to OD=1.7 because still the rest are not ready to start.
14:40 OD measurements
OD-10X dilution
Titer
Ready to transform
Time
ylic133_1
0.34
3.4
yes
14:40
ylic133_4 (1)
0.114
1.1
let’s see , should be ~2
14:40
ylic133_4 (2)
0.28
2.8
yes
14:40
Byk832
0.24
2.4
yes
14:40
100ng of pBK549 plasmid
Note: The cells after adding the transformation mix were really clumpy, so they did not properly dissolved in the transformation mix, which can hinder the transformation efficiency.
Plating all in -ura+6xADE
67.4. Results :pensive:#
-30092019 - No colonies in -URA plates , in none of the four strains used :( There is just a loan
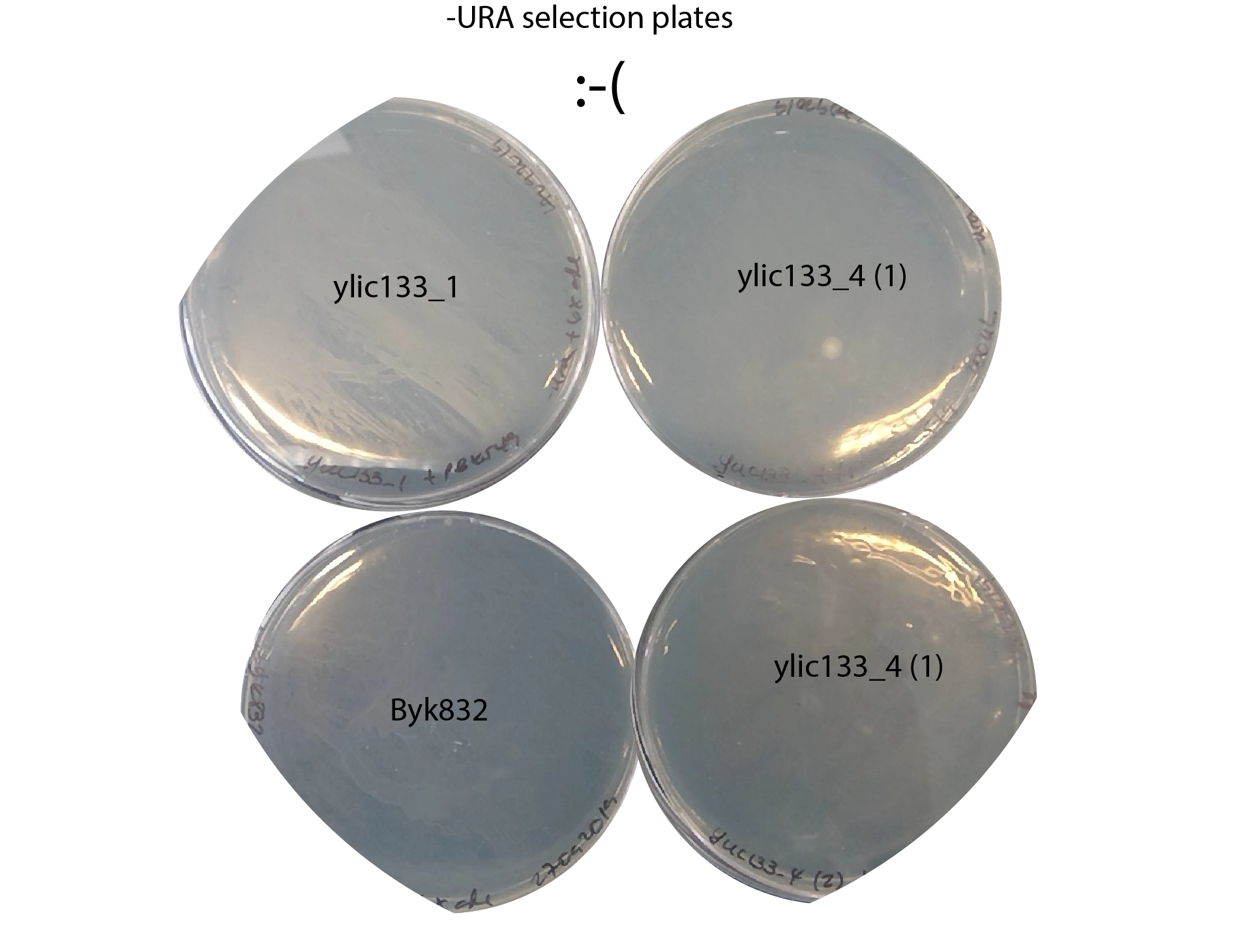
67.5. Conclusion#
Indeed, it seems the fact that the cells were clumpy place a role on the transformation. This happens to me as well with Byk832 in previous transformations, see this transformation
Repeat the transformation , and try to minimize again the time in LiAc and mix every time tha transformation mix on the cells to avoid formation of clumps.
