Title : ylic133 and ylic136 transformation with bem3d::NAT
Contents
33. Title : ylic133 and ylic136 transformation with bem3d::NAT#
33.1. Date#
14102020-
33.2. Objective#
To construct the bem3d strain to measure in SATAY.
To construct the bem3dnrp1d strain to measure in SATAY.
To be able to measure how bem1 interactors change with the genetic environment.
e=f(dbem3dnrp1dbem1)-f(dbem1)|dbem3f(dnrp1)|dbem3
To test in satay if the fitness of nrp1 in dbem3 background is equal to fitness of bem3 in dnrp1 background.
33.3. Method#
Extract the DNA from yll138a: bem3::NAT W303
15102020 (at 10:30) Inoculate some colonies from a plate in YPD+NAT media (it also has 6xADE).
16102020: DNA extraction
2mL used of dense culture
Do a PCR with primerset: bem3-upstream/bem3-downstream
Use 1ul of DNA for PCR (due to extremely low concentration of DNA , the PCR product was really faint, I used effectively 0.8ng as template.)
Transform ylic133 and ylic136 with the PCR product.
33.4. Results#
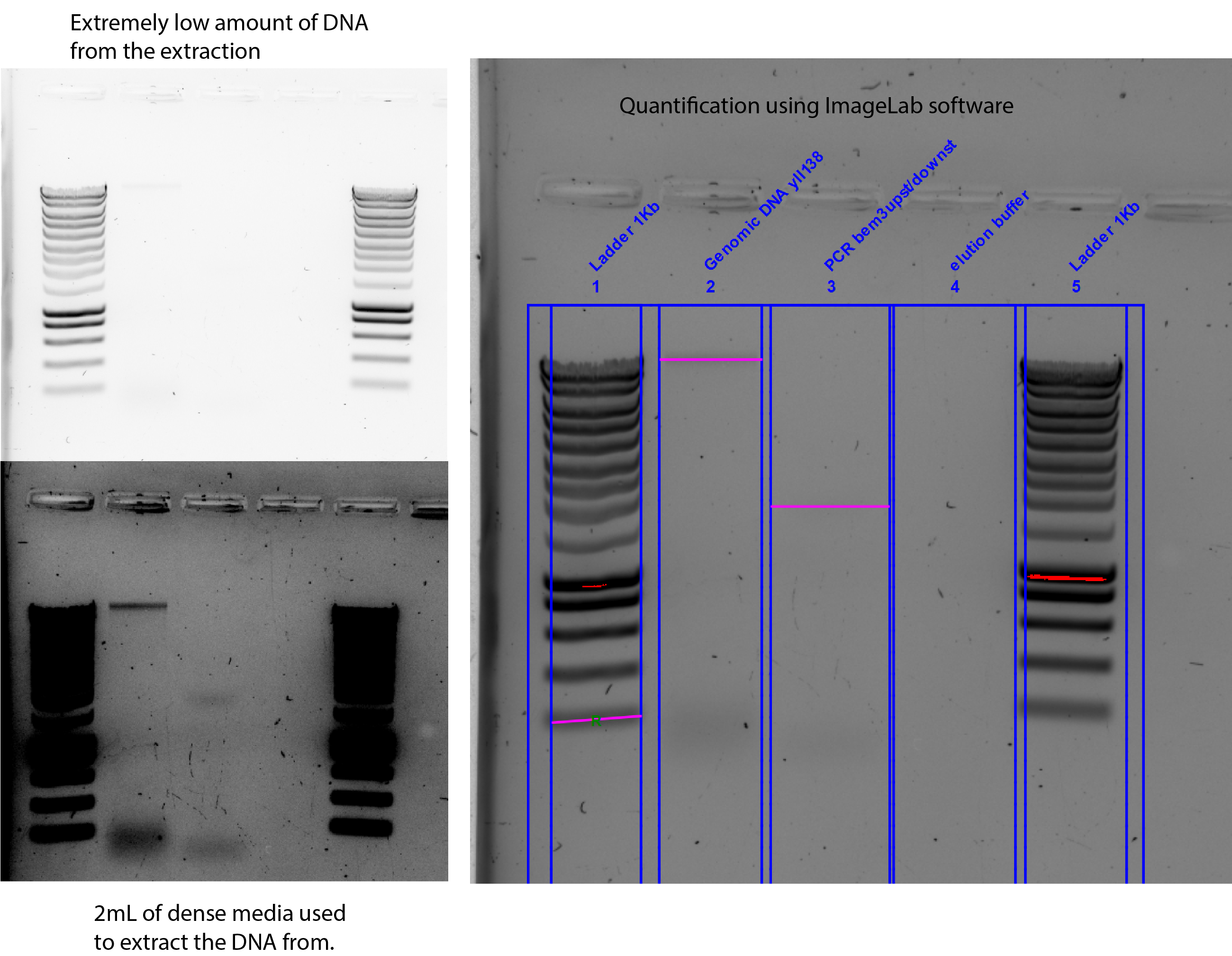
33.4.1. DNA Quantification on gel, using ImageLab software from BioRad#
reference band: 20ng
Lane2= 0.15 x reference band=3ng band
DNA concentration = 3ng/5uL=0.8ng/ul extremely low
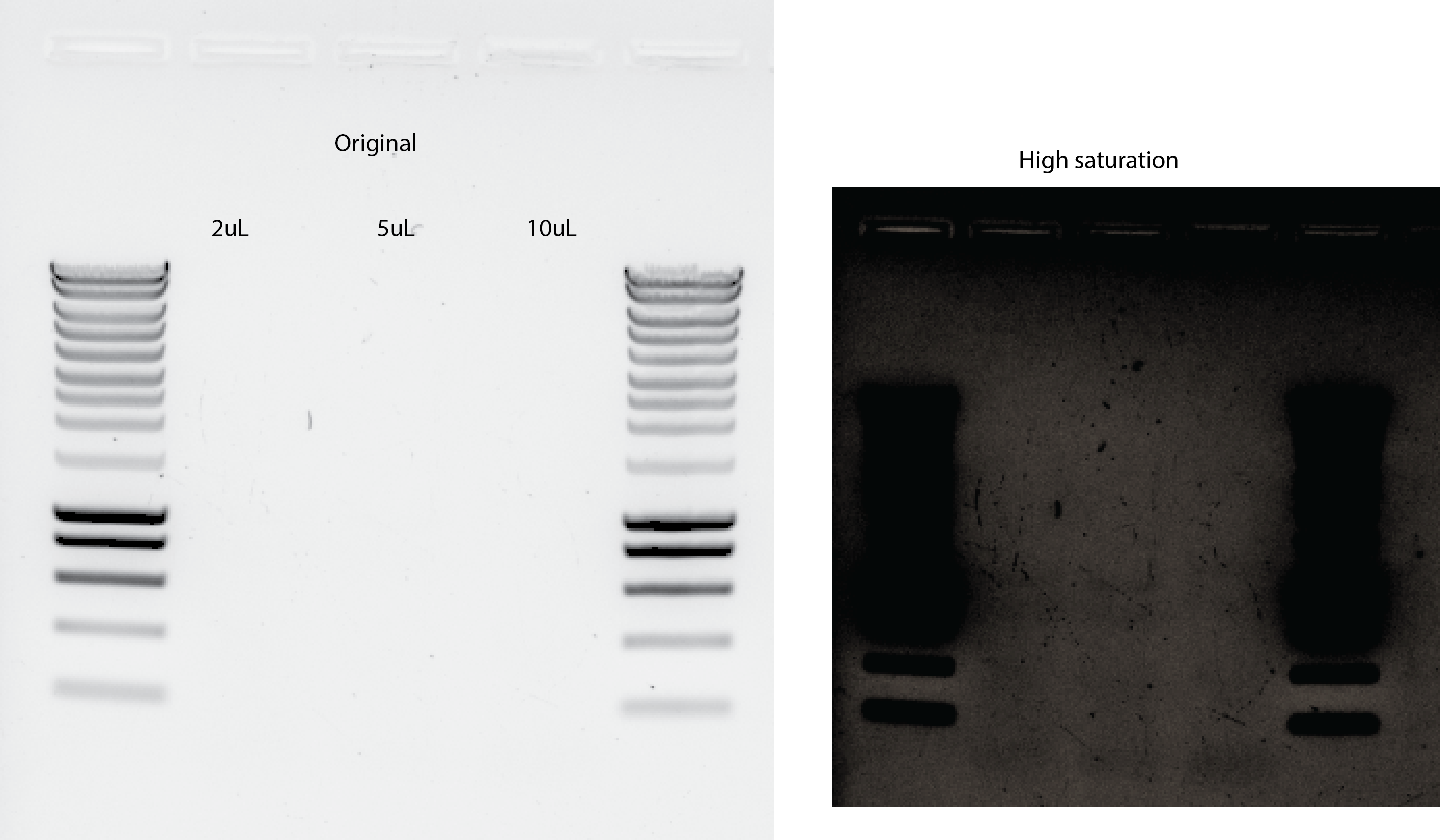
33.4.2. New PCR with more DNA :( 😭#
PCR with 2uL, 5uL and 10uL of DNA template.
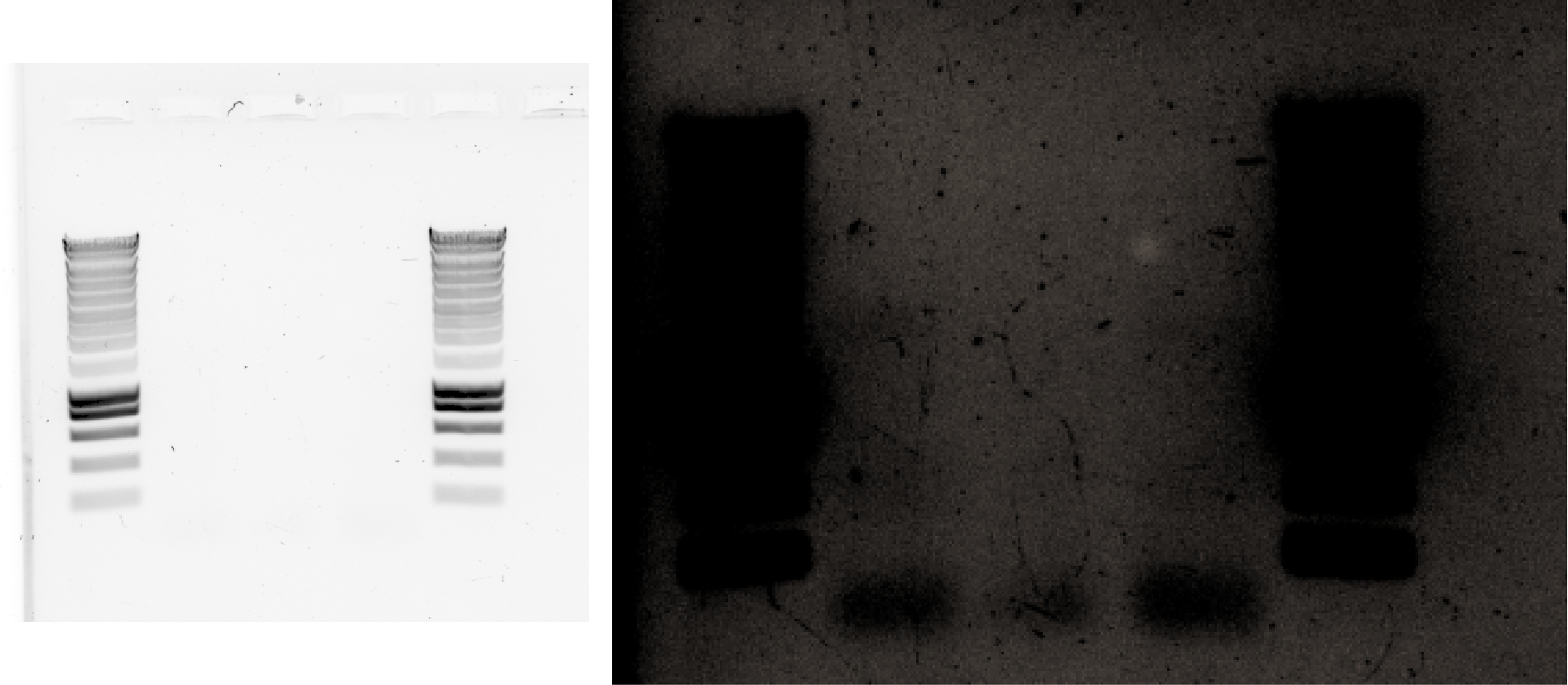
33.4.3. Try lowering the annealing temperature😭#
PCR with 2uL, 5uL and 10uL of DNA template, using T=55C as annealing temperature.
The used primers have a melting temperature of: 57C and 61C respectively. From this website : they say that The optimal annealing temperature should be determined empirically, but it is typically lower than the primers’ Tm by approximately 5°C to 10°C.
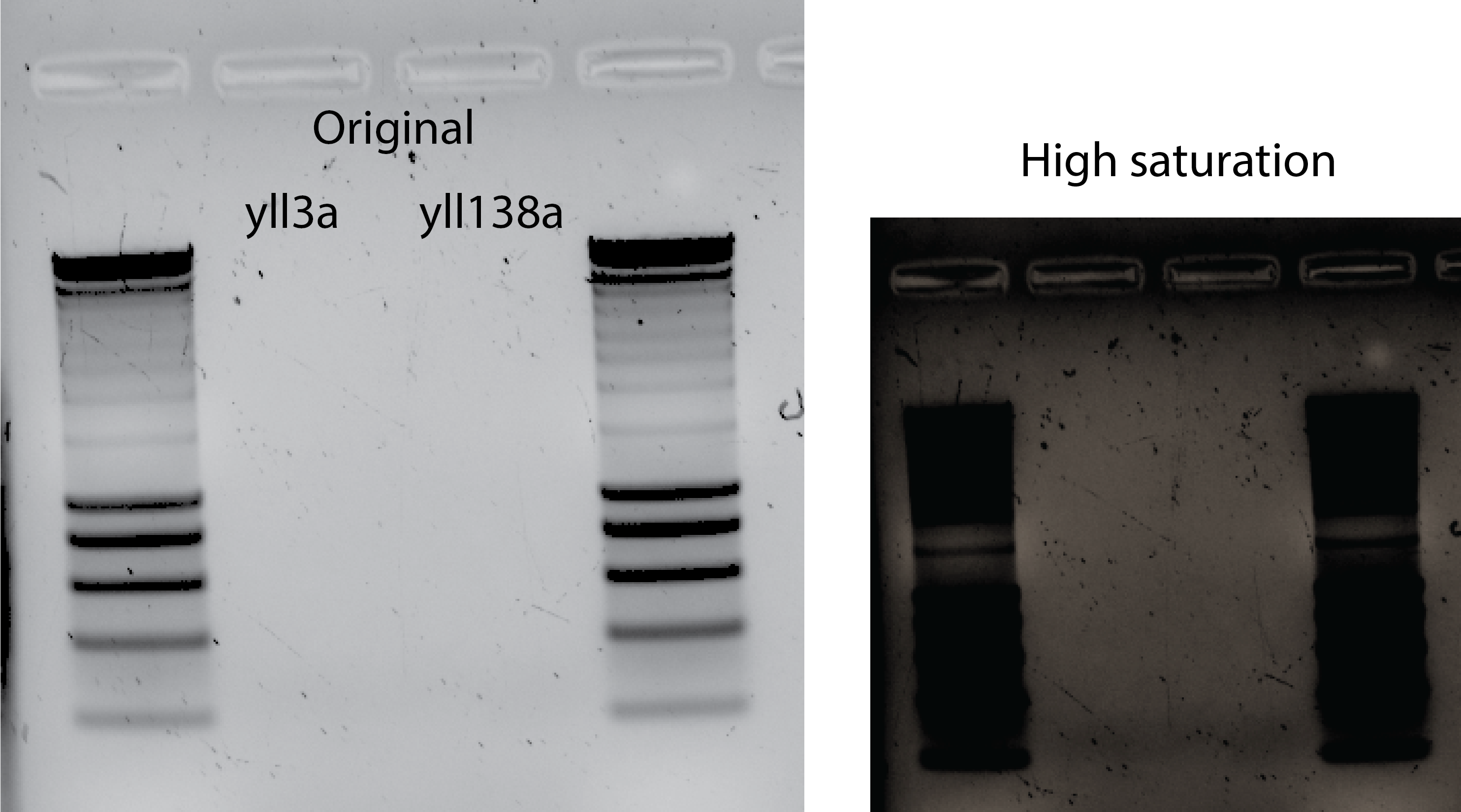
33.4.4. Try a positive control in yll3a to test the primers😭#
PCR with yll3a and 36.5uL of yll138 DNA template.
33.4.5. Try different primers (oLL401/oLL402)😭#
New DNA extraction using 10mL of cell culture
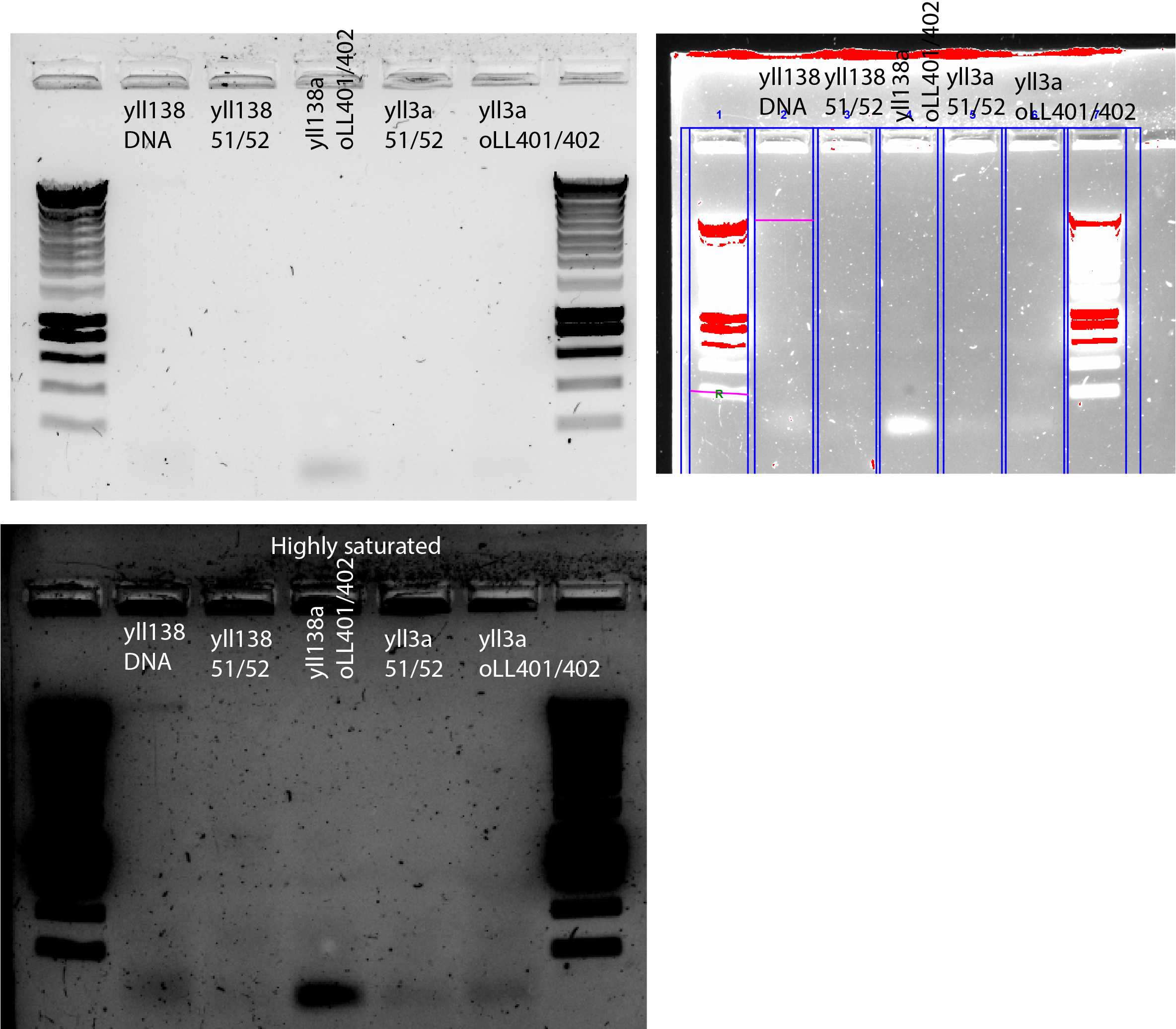
Concentration given by BioRad taking the 20ng band as reference : ~ 9ng. The loaded DNA was 5X diluted. I got 0.09* I(20ng) *5=9ng/uL
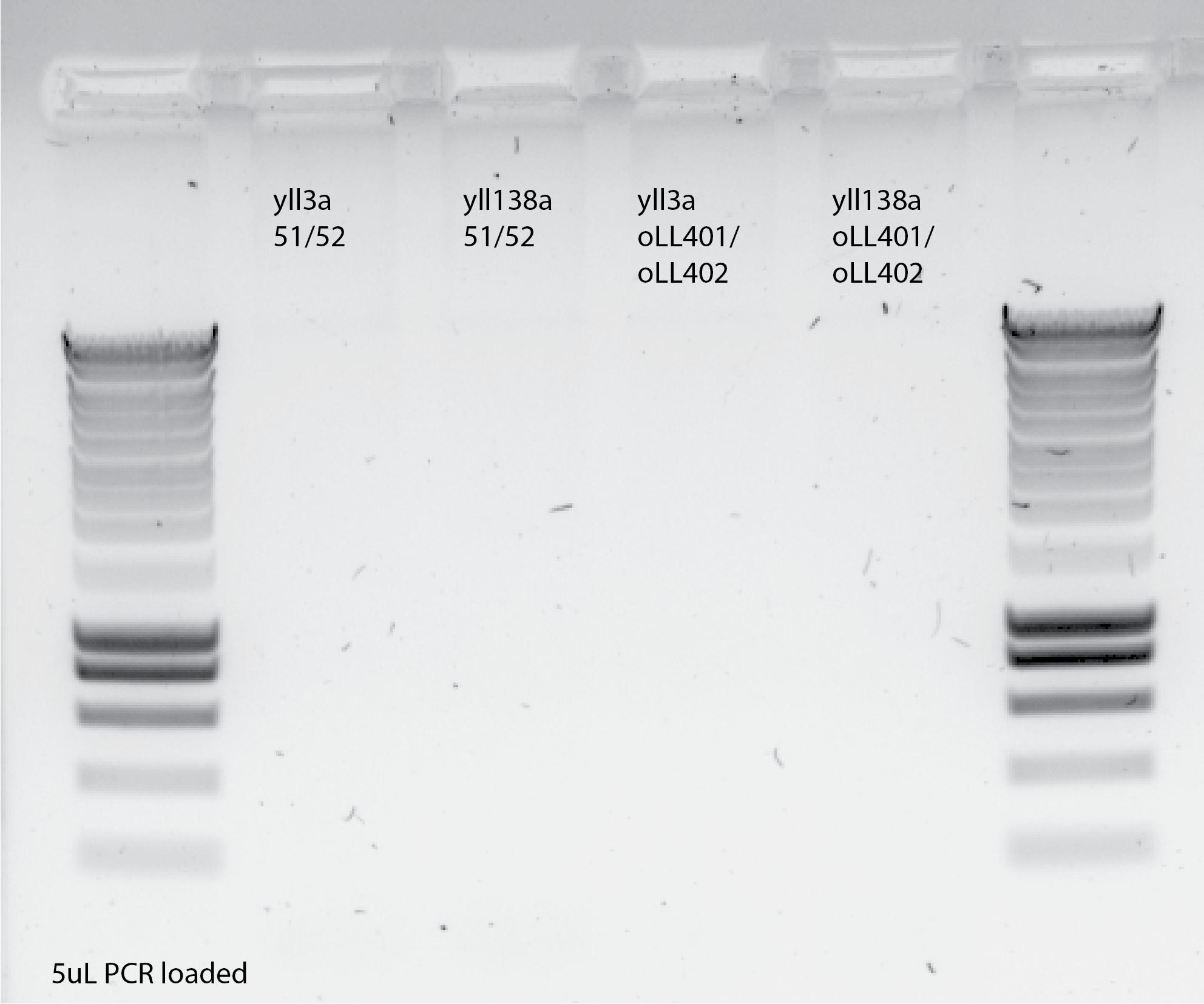
33.4.6. Try Wessel PCR protocol with oLL primers 😭#
Protocol:
3mins in 98C
30X(30s in 98C, 30s in 65C, 1min in 72C)
12 mins in 72C
4C on hold
In my previous protocol I used
3mins in 95C
35X(30s in 95C, 30s in 55C, 1min in 72C)
12 mins in 72C
12C on hold
 {width=50%}
{width=50%}
33.4.7. Try different set of primers that have worked for me before#
From this experiment I to check the presence and deletion of bem2 in different strains , I can use:
primers 41/47 that will bind to bem2 gene and upstream from it.
Check on yll3a and yll138 template
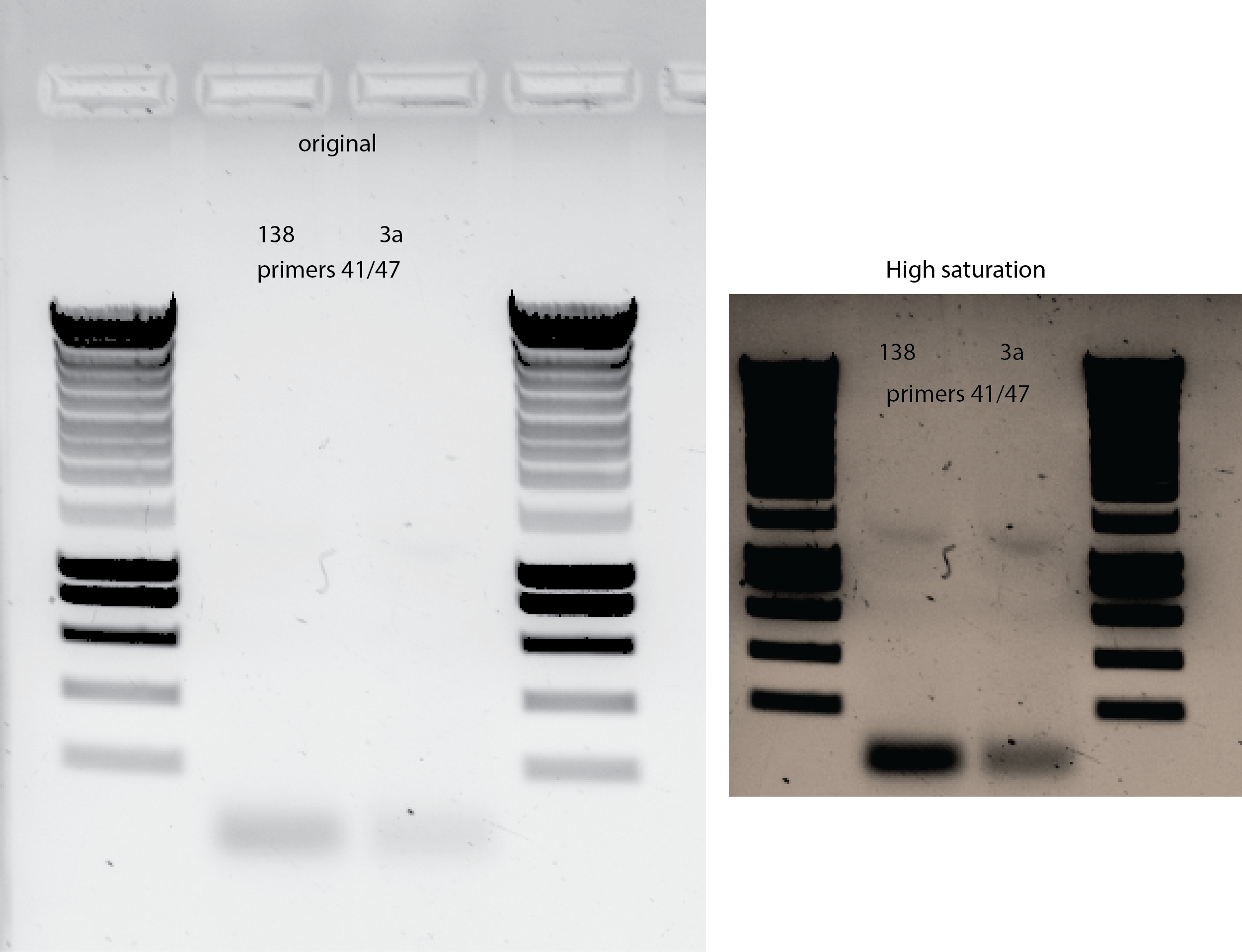
Next time , I need to MIX , VORTEX THE GENOMIC DNA BECAUSE IT WILL SEDIMENT MOST OF IT. So maybe I was taking very low amount , that is why I have this low efficiency.
33.4.8. Try mixing the gDNA before PCR#
PCR with primer 41/47 and 51/52 and vortexing first the DNA from yll3a and yll138a
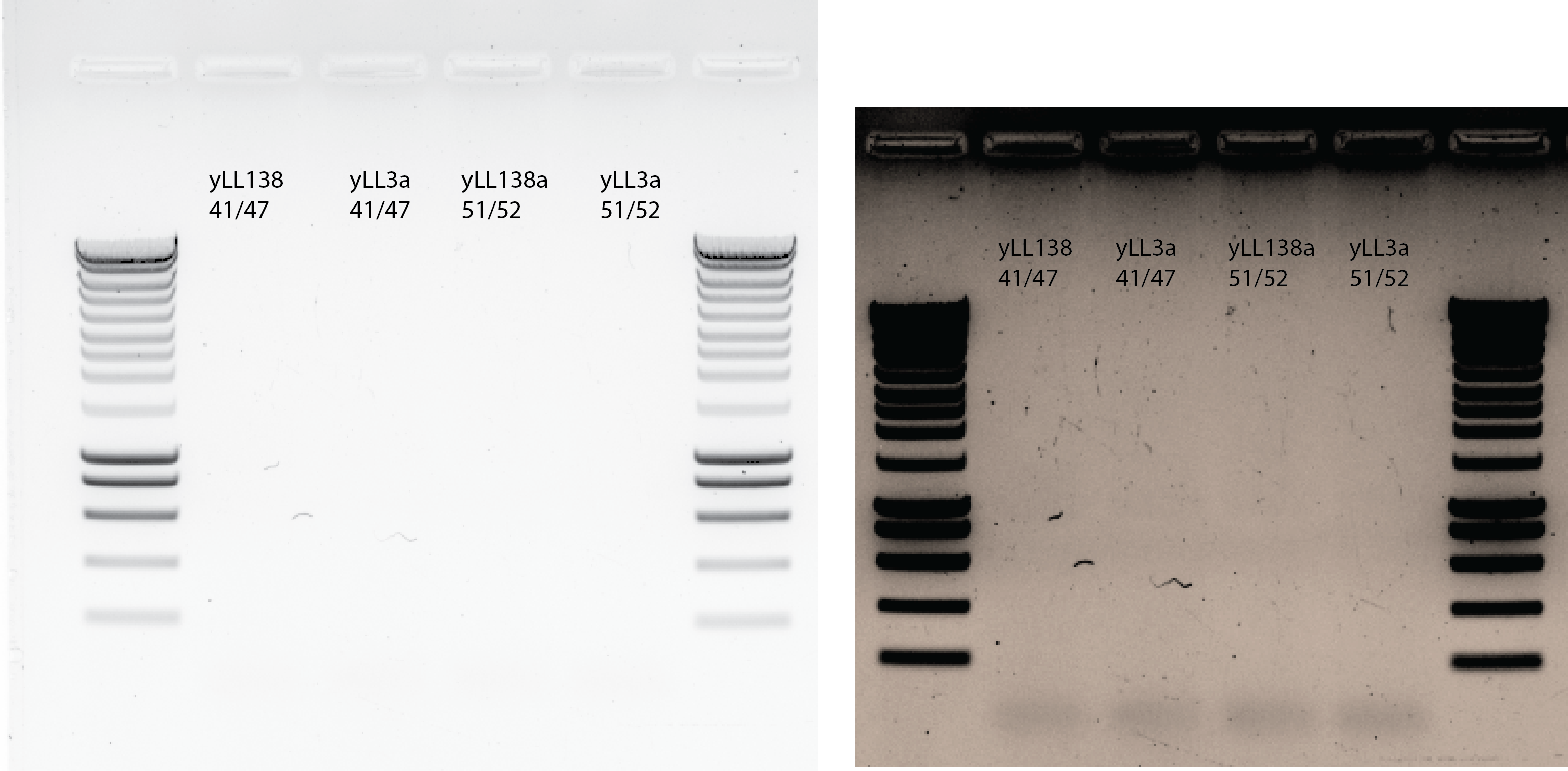
33.5. Conclusion#
Unsuccessful PCR attempts with the gDNA from yll138 and those primers. This is the PCR product I would need for transforming my ylic133 strain.
Next steps: Extract more gDNA and repeat with more details all these steps .
